Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4
- PMID: 27347908
- PMCID: PMC6273265
- DOI: 10.3390/molecules21070822
Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4
Abstract
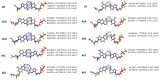
Expanded (x) and widened (y) deoxyribose nucleic acids (DNA) have an extra benzene ring incorporated either horizontally (xDNA) or vertically (yDNA) between a natural pyrimidine base and the deoxyribose, or between the 5- and 6-membered rings of a natural purine. Far-reaching applications for (x,y)DNA include nucleic acid probes and extending the natural genetic code. Since modified nucleobases must encode information that can be passed to the next generation in order to be a useful extension of the genetic code, the ability of translesion (bypass) polymerases to replicate modified bases is an active area of research. The common model bypass polymerase DNA polymerase IV (Dpo4) has been previously shown to successfully replicate and extend past a single modified nucleobase on a template DNA strand. In the current study, molecular dynamics (MD) simulations are used to evaluate the accommodation of expanded/widened nucleobases in the Dpo4 active site, providing the first structural information on the replication of (x,y)DNA. Our results indicate that the Dpo4 catalytic (palm) domain is not significantly impacted by the (x,y)DNA bases. Instead, the template strand is displaced to accommodate the increased C1'-C1' base-pair distance. The structural insights unveiled in the present work not only increase our fundamental understanding of Dpo4 replication, but also reveal the process by which Dpo4 replicates (x,y)DNA, and thereby will contribute to the optimization of high fidelity and efficient polymerases for the replication of modified nucleobases.
Keywords: DNA replication; Dpo4; bypass polymerase; expanded DNA; molecular dynamics; translesion synthesis; widened DNA; xDNA; yDNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




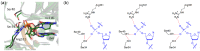



References
-
- Webb S.J. Bioinspired organic chemistry. Annu. Rep. Prog. Chem. Sect. B Org. Chem. 2006;102:377–400. doi: 10.1039/b515108m. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

