Replication-Competent Influenza A Viruses Expressing Reporter Genes
- PMID: 27347991
- PMCID: PMC4974514
- DOI: 10.3390/v8070179
Replication-Competent Influenza A Viruses Expressing Reporter Genes
Abstract
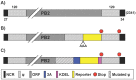
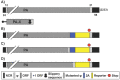
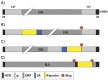
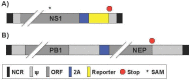
Influenza A viruses (IAV) cause annual seasonal human respiratory disease epidemics. In addition, IAV have been implicated in occasional pandemics with inordinate health and economic consequences. Studying IAV, in vitro or in vivo, requires the use of laborious secondary methodologies to identify virus-infected cells. To circumvent this requirement, replication-competent IAV expressing an easily traceable reporter protein can be used. Here we discuss the development and applications of recombinant replication-competent IAV harboring diverse fluorescent or bioluminescent reporter genes in different locations of the viral genome. These viruses have been employed for in vitro and in vivo studies, such as the screening of neutralizing antibodies or antiviral compounds, the identification of host factors involved in viral replication, cell tropism, the development of vaccines, or the assessment of viral infection dynamics. In summary, reporter-expressing, replicating-competent IAV represent a powerful tool for the study of IAV both in vitro and in vivo.
Keywords: fluorescence; luminescence; plasmid-based reverse genetics; recombinant influenza A virus; replicating-competent reporter-expressing influenza A virus; reporter genes; virus rescue approaches.
Figures



References
-
- Palese P., Shaw M.L. Orthomyxoviridae: The viruses and their replication. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., editors. Fields Virology. 5th ed. Lippincott Williams and Wilkins; Philadelphia, PA, USA: 2007.
-
- WHO . fluenza (Seasonal) Fact Sheet No. 211. WHO; Geneva, Switzerland: 2009.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

