Auxotrophic Actinobacillus pleurpneumoniae grows in multispecies biofilms without the need for nicotinamide-adenine dinucleotide (NAD) supplementation
- PMID: 27349384
- PMCID: PMC4924255
- DOI: 10.1186/s12866-016-0742-3
Auxotrophic Actinobacillus pleurpneumoniae grows in multispecies biofilms without the need for nicotinamide-adenine dinucleotide (NAD) supplementation
Abstract
Background: Actinobacillus pleuropneumoniae is the etiologic agent of porcine contagious pleuropneumonia, which causes important worldwide economic losses in the swine industry. Several respiratory tract infections are associated with biofilm formation, and A. pleuropneumoniae has the ability to form biofilms in vitro. Biofilms are structured communities of bacterial cells enclosed in a self-produced polymer matrix that are attached to an abiotic or biotic surface. Virtually all bacteria can grow as a biofilm, and multi-species biofilms are the most common form of microbial growth in nature. The goal of this study was to determine the ability of A. pleuropneumoniae to form multi-species biofilms with other bacteria frequently founded in pig farms, in the absence of pyridine compounds (nicotinamide mononucleotide [NMN], nicotinamide riboside [NR] or nicotinamide adenine dinucleotide [NAD]) that are essential for the growth of A. pleuropneumoniae.
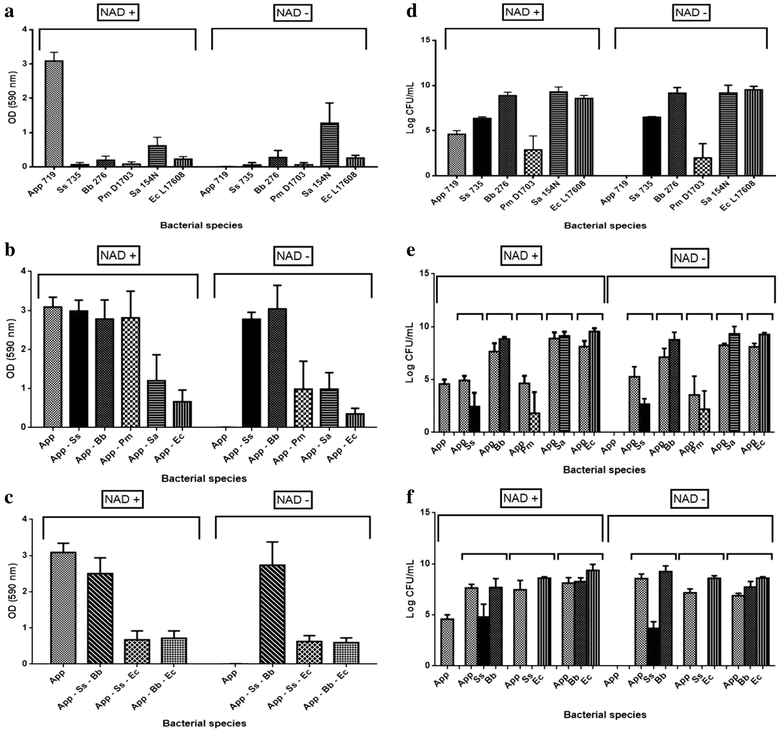
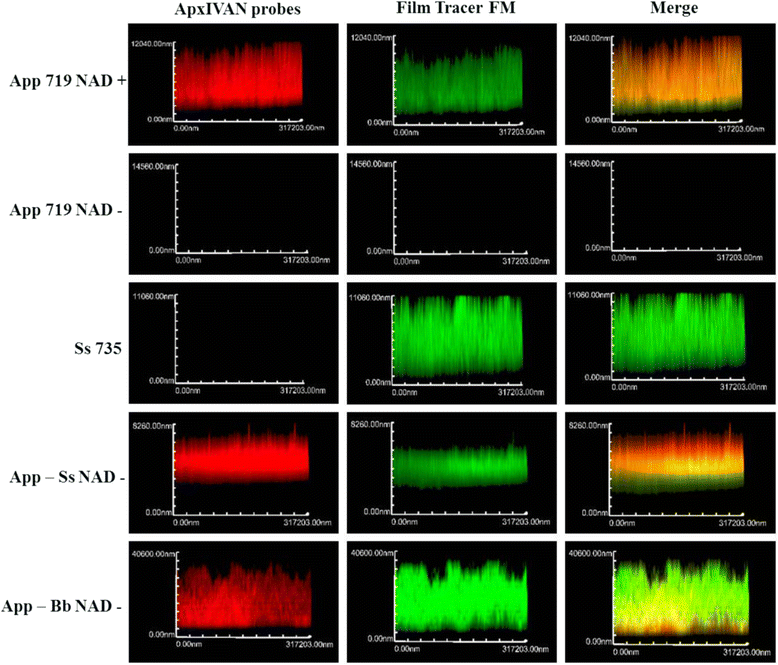
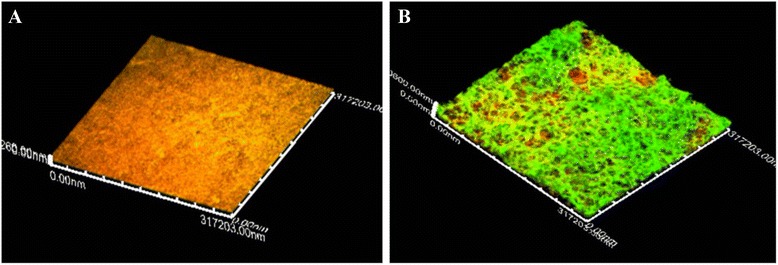
Results: For the biofilm assay, strain 719, a field isolate of A. pleuropneumoniae serovar 1, was mixed with swine isolates of Streptococcus suis, Bordetella bronchiseptica, Pasteurella multocida, Staphylococcus aureus or Escherichia coli, and deposited in 96-well microtiter plates. Based on the CFU results, A. pleuropneumoniae was able to grow with every species tested in the absence of pyridine compounds in the culture media. Interestingly, A. pleuropneumoniae was also able to form strong biofilms when mixed with S. suis, B. bronchiseptica or S. aureus. In the presence of E. coli, A. pleuropneumoniae only formed a weak biofilm. The live and dead populations, and the matrix composition of multi-species biofilms were also characterized using fluorescent markers and enzyme treatments. The results indicated that poly-N-acetyl-glucosamine remains the primary component responsible for the biofilm structure.
Conclusions: In conclusion, A. pleuropneumoniae apparently is able to satisfy the requirement of pyridine compounds through of other swine pathogens by cross-feeding, which enables A. pleuropneumoniae to grow and form multi-species biofilms.
Keywords: Actinobacillus pleuropneumoniae; Biofilms; Bordetella bronchiseptica; Escherichia coli; Pasteurella multocida; Pyridine compounds; Staphylococcus aureus; Streptococcus suis.
Figures


Similar articles
-
Sub-inhibitory concentrations of penicillin G induce biofilm formation by field isolates of Actinobacillus pleuropneumoniae.Vet Microbiol. 2015 Sep 30;179(3-4):277-86. doi: 10.1016/j.vetmic.2015.06.011. Epub 2015 Jun 24. Vet Microbiol. 2015. PMID: 26130517
-
Method to grow Actinobacillus pleuropneumoniae biofilm on a biotic surface.BMC Vet Res. 2013 Oct 20;9:213. doi: 10.1186/1746-6148-9-213. BMC Vet Res. 2013. PMID: 24139070 Free PMC article.
-
Actinobacillus pleuropneumoniae grows as aggregates in the lung of pigs: is it time to refine our in vitro biofilm assays?Microb Biotechnol. 2017 Jul;10(4):756-760. doi: 10.1111/1751-7915.12432. Epub 2016 Oct 28. Microb Biotechnol. 2017. PMID: 27790837 Free PMC article.
-
Actinobacillus pleuropneumoniae biofilms: Role in pathogenicity and potential impact for vaccination development.Anim Health Res Rev. 2018 Jun;19(1):17-30. doi: 10.1017/S146625231700010X. Epub 2017 Nov 7. Anim Health Res Rev. 2018. PMID: 29110751 Review.
-
Surface polysaccharides and iron-uptake systems of Actinobacillus pleuropneumoniae.Can J Vet Res. 2004 Apr;68(2):81-5. Can J Vet Res. 2004. PMID: 15188950 Free PMC article. Review.
Cited by
-
Incorporation of Actinobacillus pleuropneumoniae in Preformed Biofilms by Escherichia coli Isolated From Drinking Water of Swine Farms.Front Vet Sci. 2018 Aug 14;5:184. doi: 10.3389/fvets.2018.00184. eCollection 2018. Front Vet Sci. 2018. PMID: 30155471 Free PMC article.
-
Shifts in the swine nasal microbiota following Bordetella bronchiseptica challenge in a longitudinal study.Front Microbiol. 2023 Sep 29;14:1260465. doi: 10.3389/fmicb.2023.1260465. eCollection 2023. Front Microbiol. 2023. PMID: 37840723 Free PMC article.
-
Actinobacillus pleuropneumoniae Surviving on Environmental Multi-Species Biofilms in Swine Farms.Front Vet Sci. 2021 Sep 30;8:722683. doi: 10.3389/fvets.2021.722683. eCollection 2021. Front Vet Sci. 2021. PMID: 34660763 Free PMC article.
-
The morphology and metabolic changes of Actinobacillus pleuropneumoniae during its growth as a biofilm.Vet Res. 2023 May 26;54(1):42. doi: 10.1186/s13567-023-01173-x. Vet Res. 2023. PMID: 37237397 Free PMC article.
-
Effect of Simultaneous Exposure of Pigs to Streptococcus suis Serotypes 2 and 9 on Their Colonization and Transmission, and on Mortality.Pathogens. 2017 Sep 27;6(4):46. doi: 10.3390/pathogens6040046. Pathogens. 2017. PMID: 28953248 Free PMC article.
References
-
- Brockmeier S, Halbur P, Thacker, E. Porcine Respiratory Disease Complex. In: Brogden K, Guthmiller J, editors. Polymicrobial Diseases. ASM Press: American Society for Microbiology; 2002;231-58.
-
- Ramjeet M, Cox A, Hancock M, Mourez M, Labrie J, et al. Mutation in the LPS outer core biosynthesis gene, galU, affects LPS interaction with the RTX toxins ApxI and ApxII and cytolytic activity of Actinobacillus pleuropneumoniae serovar 1. Mol Microbiol. 2008;70:221–235. doi: 10.1111/j.1365-2958.2008.06409.x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

