miR-93 regulates Msk2-mediated chromatin remodelling in diabetic nephropathy
- PMID: 27350436
- PMCID: PMC4931323
- DOI: 10.1038/ncomms12076
miR-93 regulates Msk2-mediated chromatin remodelling in diabetic nephropathy
Abstract
How the kidney responds to the metabolic cues from the environment remains a central question in kidney research. This question is particularly relevant to the pathogenesis of diabetic nephropathy (DN) in which evidence suggests that metabolic events in podocytes regulate chromatin structure. Here, we show that miR-93 is a critical metabolic/epigenetic switch in the diabetic milieu linking the metabolic state to chromatin remodelling. Mice with inducible overexpression of a miR-93 transgene exclusively in podocytes exhibit significant improvements in key features of DN. We identify miR-93 as a regulator of nucleosomal dynamics in podocytes. miR-93 has a critical role in chromatin reorganization and progression of DN by modulating its target Msk2, a histone kinase, and its substrate H3S10. These findings implicate a central role for miR-93 in high glucose-induced chromatin remodelling in the kidney, and provide evidence for a previously unrecognized role for Msk2 as a target for DN therapy.
Figures

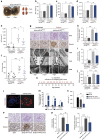


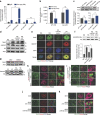
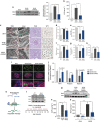
Comment in
-
Diabetic nephropathy: Chromatin remodelling in diabetic nephropathy.Nat Rev Nephrol. 2016 Jul 18;12(8):444. doi: 10.1038/nrneph.2016.106. Nat Rev Nephrol. 2016. PMID: 27425389 No abstract available.
References
-
- U.S. Renal Data System. USRDS 2013 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. (National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2013).
-
- van Rooij E. et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007). - PubMed
-
- Trajkovski M. et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

