Utilizing mutual information for detecting rare and common variants associated with a categorical trait
- PMID: 27350900
- PMCID: PMC4918222
- DOI: 10.7717/peerj.2139
Utilizing mutual information for detecting rare and common variants associated with a categorical trait
Abstract
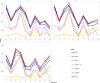
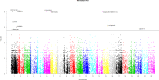
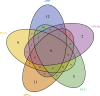
Background. Genome-wide association studies have succeeded in detecting novel common variants which associate with complex diseases. As a result of the fast changes in next generation sequencing technology, a large number of sequencing data are generated, which offers great opportunities to identify rare variants that could explain a larger proportion of missing heritability. Many effective and powerful methods are proposed, although they are usually limited to continuous, dichotomous or ordinal traits. Notice that traits having nominal categorical features are commonly observed in complex diseases, especially in mental disorders, which motivates the incorporation of the characteristics of the categorical trait into association studies with rare and common variants. Methods. We construct two simple and intuitive nonparametric tests, MIT and aMIT, based on mutual information for detecting association between genetic variants in a gene or region and a categorical trait. MIT and aMIT can gauge the difference among the distributions of rare and common variants across a region given every categorical trait value. If there is little association between variants and a categorical trait, MIT or aMIT approximately equals zero. The larger the difference in distributions, the greater values MIT and aMIT have. Therefore, MIT and aMIT have the potential for detecting functional variants. Results.We checked the validity of proposed statistics and compared them to the existing ones through extensive simulation studies with varied combinations of the numbers of variants of rare causal, rare non-causal, common causal, and common non-causal, deleterious and protective, various minor allele frequencies and different levels of linkage disequilibrium. The results show our methods have higher statistical power than conventional ones, including the likelihood based score test, in most cases: (1) there are multiple genetic variants in a gene or region; (2) both protective and deleterious variants are present; (3) there exist rare and common variants; and (4) more than half of the variants are neutral. The proposed tests are applied to the data from Collaborative Studies on Genetics of Alcoholism, and a competent performance is exhibited therein. Discussion. As a complementary to the existing methods mainly focusing on quantitative traits, this study provides the nonparametric tests MIT and aMIT for detecting variants associated with categorical trait. Furthermore, we plan to investigate the association between rare variants and multiple categorical traits.
Keywords: Association analysis; Categorical trait; Mutual information; Next generation sequencing data; Rare variant.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
Similar articles
-
Weighted selective collapsing strategy for detecting rare and common variants in genetic association study.BMC Genet. 2012 Feb 6;13:7. doi: 10.1186/1471-2156-13-7. BMC Genet. 2012. PMID: 22309429 Free PMC article.
-
Detecting multiple variants associated with disease based on sequencing data of case-parent trios.J Hum Genet. 2016 Oct;61(10):851-860. doi: 10.1038/jhg.2016.63. Epub 2016 Jun 9. J Hum Genet. 2016. PMID: 27278787
-
Association detection between ordinal trait and rare variants based on adaptive combination of P values.J Hum Genet. 2018 Jan;63(1):37-45. doi: 10.1038/s10038-017-0354-2. Epub 2017 Nov 7. J Hum Genet. 2018. PMID: 29215083
-
Identifying rare variants associated with complex traits via sequencing.Curr Protoc Hum Genet. 2013 Jul;Chapter 1:Unit 1.26. doi: 10.1002/0471142905.hg0126s78. Curr Protoc Hum Genet. 2013. PMID: 23853079 Free PMC article. Review.
-
Linkage analysis in the next-generation sequencing era.Hum Hered. 2011;72(4):228-36. doi: 10.1159/000334381. Epub 2011 Dec 23. Hum Hered. 2011. PMID: 22189465 Free PMC article. Review.
Cited by
-
MIDESP: Mutual Information-Based Detection of Epistatic SNP Pairs for Qualitative and Quantitative Phenotypes.Biology (Basel). 2021 Sep 16;10(9):921. doi: 10.3390/biology10090921. Biology (Basel). 2021. PMID: 34571798 Free PMC article.
-
Improving power of genome-wide association studies via transforming ordinal phenotypes into continuous phenotypes.Front Plant Sci. 2023 Nov 2;14:1247181. doi: 10.3389/fpls.2023.1247181. eCollection 2023. Front Plant Sci. 2023. PMID: 38023883 Free PMC article.
-
Rare variant association analysis in case-parents studies by allowing for missing parental genotypes.BMC Genet. 2018 Jan 15;19(1):7. doi: 10.1186/s12863-018-0597-8. BMC Genet. 2018. PMID: 29334894 Free PMC article.
-
Epistatic evidence for gender-dependant slow neurotransmission signalling in substance use disorders: PPP1R12B versus PPP1R1B.EBioMedicine. 2020 Nov;61:103066. doi: 10.1016/j.ebiom.2020.103066. Epub 2020 Oct 21. EBioMedicine. 2020. PMID: 33096475 Free PMC article.
References
-
- Agresti A. Categorical data analysis. Wiley; Hoboken: 2012.
-
- American Psychiatric Association Diagnostic and statistical manual of mental disorders. American Psychiatric Association; Washington, D.C: 1994.
-
- Brunel H, Gallardo-Chacón JJ, Buil A, Vallverdú M, Soria JM, Caminal P, Perera A. MISS: a non-linear methodology based on mutual information for genetic association studies in both population and sib-pairs analysis. Bioinformatics. 2010;26(15):1811–1818. doi: 10.1093/bioinformatics/btq273. - DOI - PubMed
-
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JJ, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of Studies on Alcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

