Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways
- PMID: 27351335
- PMCID: PMC5026608
- DOI: 10.1021/acschembio.6b00348
Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways
Abstract
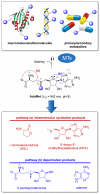
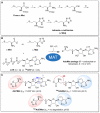
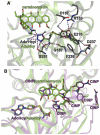
S-adenosyl-l-methionine (AdoMet) is an essential enzyme cosubstrate in fundamental biology with an expanding range of biocatalytic and therapeutic applications. We report the design, synthesis, and evaluation of stable, functional AdoMet isosteres that are resistant to the primary contributors to AdoMet degradation (depurination, intramolecular cyclization, and sulfonium epimerization). Corresponding biochemical and structural studies demonstrate the AdoMet surrogates to serve as competent enzyme cosubstrates and to bind a prototypical class I model methyltransferase (DnrK) in a manner nearly identical to AdoMet. Given this conservation in function and molecular recognition, the isosteres presented are anticipated to serve as useful surrogates in other AdoMet-dependent processes and may also be resistant to, and/or potentially even inhibit, other therapeutically relevant AdoMet-dependent metabolic transformations (such as the validated drug target AdoMet decarboxylase). This work also highlights the ability of the prototypical class I model methyltransferase DnrK to accept non-native surrogate acceptors as an enabling feature of a new high-throughput methyltransferase assay.
Figures

Similar articles
-
AdoMet analog synthesis and utilization: current state of the art.Curr Opin Biotechnol. 2016 Dec;42:189-197. doi: 10.1016/j.copbio.2016.07.005. Epub 2016 Aug 6. Curr Opin Biotechnol. 2016. PMID: 27506965 Free PMC article. Review.
-
Synthesis of S-Adenosyl-L-Methionine Analogs with Extended Transferable Groups for Methyltransferase-Directed Labeling of DNA and RNA.Curr Protoc. 2023 Jun;3(6):e799. doi: 10.1002/cpz1.799. Curr Protoc. 2023. PMID: 37327316
-
Synthesis of S-Adenosyl-L-Methionine Analogs with Extended Transferable Groups for Methyltransferase-Directed Labeling of DNA and RNA.Curr Protoc Nucleic Acid Chem. 2016 Mar 1;64:1.36.1-1.36.13. doi: 10.1002/0471142700.nc0136s64. Curr Protoc Nucleic Acid Chem. 2016. PMID: 26967468 Free PMC article.
-
Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.J Am Chem Soc. 2013 Oct 16;135(41):15536-48. doi: 10.1021/ja407140k. Epub 2013 Oct 7. J Am Chem Soc. 2013. PMID: 24093804
-
S-Adenosyl-L-methionine: beyond the universal methyl group donor.Phytochemistry. 2006 Aug;67(15):1686-98. doi: 10.1016/j.phytochem.2006.04.019. Phytochemistry. 2006. PMID: 16766004 Review.
Cited by
-
Enzymkatalysierte späte Modifizierungen: Besser spät als nie.Angew Chem Weinheim Bergstr Ger. 2021 Jul 26;133(31):16962-16993. doi: 10.1002/ange.202014931. Epub 2021 Mar 8. Angew Chem Weinheim Bergstr Ger. 2021. PMID: 38505660 Free PMC article. Review.
-
Biochemical and Structural Studies of the Carminomycin 4-O-Methyltransferase DnrK.J Nat Prod. 2024 Apr 26;87(4):798-809. doi: 10.1021/acs.jnatprod.3c00947. Epub 2024 Feb 27. J Nat Prod. 2024. PMID: 38412432 Free PMC article.
-
Organophosphorus S-adenosyl-L-methionine mimetics: synthesis, stability, and substrate properties.Front Chem. 2024 Aug 1;12:1448747. doi: 10.3389/fchem.2024.1448747. eCollection 2024. Front Chem. 2024. PMID: 39148665 Free PMC article.
-
Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.ACS Chem Biol. 2020 Mar 20;15(3):695-705. doi: 10.1021/acschembio.9b00943. Epub 2020 Mar 3. ACS Chem Biol. 2020. PMID: 32091873 Free PMC article.
-
Investigation of a squaramide motif as a bioisostere of the amino-acid group of S-adenosyl-L-methionine and its functional impact on RNA methylation.Commun Chem. 2025 Aug 12;8(1):244. doi: 10.1038/s42004-025-01627-7. Commun Chem. 2025. PMID: 40797095 Free PMC article.
References
-
- Struck A-W, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. ChemBioChem. 2012;13:2642–2655. - PubMed
-
- Wessjohann LA, Keim J, Weigel B, Dippe M. Alkylating enzymes. Curr. Opin. Chem. Biol. 2013;17:229–235. - PubMed
-
- Vance DE. Phospholipid methylation in mammals: From biochemistry to physiological function. Biochim. Biophys. Acta, Biomembr. 2014;1838:1477–1487. - PubMed
-
- Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 2012;29:1238–1250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

