Expression and purification of the modification-dependent restriction enzyme BisI and its homologous enzymes
- PMID: 27353146
- PMCID: PMC4926085
- DOI: 10.1038/srep28579
Expression and purification of the modification-dependent restriction enzyme BisI and its homologous enzymes
Abstract
The methylation-dependent restriction endonuclease (REase) BisI (G(m5)C ↓ NGC) is found in Bacillus subtilis T30. We expressed and purified the BisI endonuclease and 34 BisI homologs identified in bacterial genomes. 23 of these BisI homologs are active based on digestion of (m5)C-modified substrates. Two major specificities were found among these BisI family enzymes: Group I enzymes cut GCNGC containing two to four (m5)C in the two strands, or hemi-methylated sites containing two (m5)C in one strand; Group II enzymes only cut GCNGC sites containing three to four (m5)C, while one enzyme requires all four cytosines to be modified for cleavage. Another homolog, Esp638I cleaves GCS ↓ SGC (relaxed specificity RCN ↓ NGY, containing at least four (m5)C). Two BisI homologs show degenerate specificity cleaving unmodified DNA. Many homologs are small proteins ranging from 150 to 190 amino acid (aa) residues, but some homologs associated with mobile genetic elements are larger and contain an extra C-terminal domain. More than 156 BisI homologs are found in >60 bacterial genera, indicating that these enzymes are widespread in bacteria. They may play an important biological function in restricting pre-modified phage DNA.
Conflict of interest statement
One or more of the enzymes described in this manuscript may become commercial products of New England Biolabs, Inc. BisI endonuclease is a product of SibEnzyme Ltd.
Figures
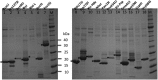




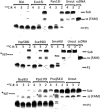
References
-
- Lacks S. & Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J. Biol. Chem. 250, 4060–4066 (1975). - PubMed
-
- Chmuzh E. V. et al.. Restriction endonuclease Bis I from Bacillus subtilis T30 recognizes methylated sequence 5′-G(m5C)↓NGC-3′ Biotechnologia (Russia) 3, 22–26 (2005).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

