Blue light-mediated transcriptional activation and repression of gene expression in bacteria
- PMID: 27353329
- PMCID: PMC5001607
- DOI: 10.1093/nar/gkw548
Blue light-mediated transcriptional activation and repression of gene expression in bacteria
Abstract
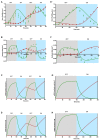
Light-regulated modules offer unprecedented new ways to control cellular behavior in precise spatial and temporal resolution. The availability of such tools may dramatically accelerate the progression of synthetic biology applications. Nonetheless, current optogenetic toolbox of prokaryotes has potential issues such as lack of rapid and switchable control, less portable, low dynamic expression and limited parts. To address these shortcomings, we have engineered a novel bidirectional promoter system for Escherichia coli that can be induced or repressed rapidly and reversibly using the blue light dependent DNA-binding protein EL222. We demonstrated that by modulating the dosage of light pulses or intensity we could control the level of gene expression precisely. We show that both light-inducible and repressible system can function in parallel with high spatial precision in a single cell and can be switched stably between ON- and OFF-states by repetitive pulses of blue light. In addition, the light-inducible and repressible expression kinetics were quantitatively analysed using a mathematical model. We further apply the system, for the first time, to optogenetically synchronize two receiver cells performing different logic behaviors over time using blue light as a molecular clock signal. Overall, our modular approach layers a transformative platform for next-generation light-controllable synthetic biology systems in prokaryotes.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Lenz P., Søgaard-Andersen L. Temporal and spatial oscillations in bacteria. Nat. Rev. Micro. 2011;9:565–577. - PubMed
-
- Bacchus W., Fussenegger M. The use of light for engineered control and reprogramming of cellular functions. Curr. Opin. Biotechnol. 2012;23:695–702. - PubMed
-
- Muller K., Weber W. Optogenetic tools for mammalian systems. Mol. Biosyst. 2013;9:596–608. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

