Plasmid Dynamics in KPC-Positive Klebsiella pneumoniae during Long-Term Patient Colonization
- PMID: 27353756
- PMCID: PMC4937214
- DOI: 10.1128/mBio.00742-16
Plasmid Dynamics in KPC-Positive Klebsiella pneumoniae during Long-Term Patient Colonization
Abstract
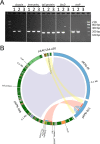
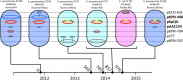
Carbapenem-resistant Klebsiella pneumoniae strains are formidable hospital pathogens that pose a serious threat to patients around the globe due to a rising incidence in health care facilities, high mortality rates associated with infection, and potential to spread antibiotic resistance to other bacterial species, such as Escherichia coli Over 6 months in 2011, 17 patients at the National Institutes of Health (NIH) Clinical Center became colonized with a highly virulent, transmissible carbapenem-resistant strain of K. pneumoniae Our real-time genomic sequencing tracked patient-to-patient routes of transmission and informed epidemiologists' actions to monitor and control this outbreak. Two of these patients remained colonized with carbapenemase-producing organisms for at least 2 to 4 years, providing the opportunity to undertake a focused genomic study of long-term colonization with antibiotic-resistant bacteria. Whole-genome sequencing studies shed light on the underlying complex microbial colonization, including mixed or evolving bacterial populations and gain or loss of plasmids. Isolates from NIH patient 15 showed complex plasmid rearrangements, leaving the chromosome and the blaKPC-carrying plasmid intact but rearranging the two other plasmids of this outbreak strain. NIH patient 16 has shown continuous colonization with blaKPC-positive organisms across multiple time points spanning 2011 to 2015. Genomic studies defined a complex pattern of succession and plasmid transmission across two different K. pneumoniae sequence types and an E. coli isolate. These findings demonstrate the utility of genomic methods for understanding strain succession, genome plasticity, and long-term carriage of antibiotic-resistant organisms.
Importance: In 2011, the NIH Clinical Center had a nosocomial outbreak involving 19 patients who became colonized or infected with blaKPC-positive Klebsiella pneumoniae Patients who have intestinal colonization with blaKPC-positive K. pneumoniae are at risk for developing infections that are difficult or nearly impossible to treat with existing antibiotic options. Two of those patients remained colonized with blaKPC-positive Klebsiella pneumoniae for over a year, leading to the initiation of a detailed genomic analysis exploring mixed colonization, plasmid recombination, and plasmid diversification. Whole-genome sequence analysis identified a variety of changes, both subtle and large, in the blaKPC-positive organisms. Long-term colonization of patients with blaKPC-positive Klebsiella pneumoniae creates new opportunities for horizontal gene transfer of plasmids encoding antibiotic resistance genes and poses complications for the delivery of health care.
Copyright © 2016 Conlan et al.
Figures


Similar articles
-
Clinical and Molecular Description of a High-Copy IncQ1 KPC-2 Plasmid Harbored by the International ST15 Klebsiella pneumoniae Clone.mSphere. 2020 Oct 7;5(5):e00756-20. doi: 10.1128/mSphere.00756-20. mSphere. 2020. PMID: 33028683 Free PMC article.
-
Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission.J Antimicrob Chemother. 2017 Nov 1;72(11):3025-3034. doi: 10.1093/jac/dkx264. J Antimicrob Chemother. 2017. PMID: 28961793 Free PMC article.
-
Outbreak of KPC-2-Producing Klebsiella pneumoniae ST76 Isolates in an Intensive Care Unit and Neurosurgery Unit.Microb Drug Resist. 2020 Sep;26(9):1009-1018. doi: 10.1089/mdr.2019.0363. Epub 2020 Mar 4. Microb Drug Resist. 2020. PMID: 32150494
-
Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding.Trends Microbiol. 2014 Dec;22(12):686-96. doi: 10.1016/j.tim.2014.09.003. Epub 2014 Oct 7. Trends Microbiol. 2014. PMID: 25304194 Free PMC article. Review.
-
Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria.South Med J. 2011 Jan;104(1):40-5. doi: 10.1097/SMJ.0b013e3181fd7d5a. South Med J. 2011. PMID: 21119555 Free PMC article. Review.
Cited by
-
Transfer of a bla CTX-M-1-carrying plasmid between different Escherichia coli strains within the human gut explored by whole genome sequencing analyses.Sci Rep. 2018 Jan 10;8(1):280. doi: 10.1038/s41598-017-18659-2. Sci Rep. 2018. PMID: 29321570 Free PMC article.
-
Multiscale Evolutionary Dynamics of Host-Associated Microbiomes.Cell. 2018 Mar 8;172(6):1216-1227. doi: 10.1016/j.cell.2018.02.015. Cell. 2018. PMID: 29522743 Free PMC article. Review.
-
Reduced Fitness Costs of mcr-1.2 Compared to Mutated pmrB in Isogenic Colistin-Resistant KPC-3-Producing Klebsiella pneumoniae.mSphere. 2019 Nov 6;4(6):e00551-19. doi: 10.1128/mSphere.00551-19. mSphere. 2019. PMID: 31694895 Free PMC article.
-
Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae.PLoS Genet. 2019 Apr 15;15(4):e1008114. doi: 10.1371/journal.pgen.1008114. eCollection 2019 Apr. PLoS Genet. 2019. PMID: 30986243 Free PMC article.
-
Coexistence of the blaNDM-1-carrying plasmid pWLK-NDM and the blaKPC-2-carrying plasmid pWLK-KPC in a Raoultella ornithinolytica isolate.Sci Rep. 2020 Feb 11;10(1):2360. doi: 10.1038/s41598-020-59341-4. Sci Rep. 2020. PMID: 32047243 Free PMC article.
References
-
- Davies SC. 2011. Infections and the rise of antimicrobial resistance, vol 2. Annual Report of the Chief Medical Officer, England. United Kingdom Department of Health, London, England. - PubMed
-
- President’s Council of Advisors on Science and Technology 2014. Report to the President on combating antibiotic resistance. Executive Office of the President; https://www.whitehouse.gov/sites/default/files/microsites/ostp/PCAST/pca....
-
- Tavares CP, Pereira PS, de Andrade Marques E, Faria C Jr, de Souza MDPAH, de Almeida R, Alves CDFM, Asensi MD, Carvalho-Assef AP. 2015. Molecular epidemiology of KPC-2-producing Enterobacteriaceae (non-Klebsiella pneumoniae) isolated from Brazil. Diagn Microbiol Infect Dis 82:326–330. doi:10.1016/j.diagmicrobio.2015.04.002. - DOI - PubMed
-
- Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai Y-C, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi:10.1126/scitranslmed.3009845. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

