NMR Characterization of Information Flow and Allosteric Communities in the MAP Kinase p38γ
- PMID: 27353957
- PMCID: PMC4926091
- DOI: 10.1038/srep28655
NMR Characterization of Information Flow and Allosteric Communities in the MAP Kinase p38γ
Abstract
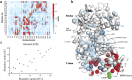
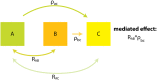
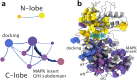
The intramolecular network structure of a protein provides valuable insights into allosteric sites and communication pathways. However, a straightforward method to comprehensively map and characterize these pathways is not currently available. Here we present an approach to characterize intramolecular network structure using NMR chemical shift perturbations. We apply the method to the mitogen activated protein kinase (MAPK) p38γ. p38γ contains allosteric sites that are conserved among eukaryotic kinases as well as unique to the MAPK family. How these regulatory sites communicate with catalytic residues is not well understood. Using our method, we observe and characterize for the first time information flux between regulatory sites through a conserved kinase infrastructure. This network is accessed, reinforced, and broken in various states of p38γ, reflecting the functional state of the protein. We demonstrate that the approach detects critical junctions in the network corresponding to biologically significant allosteric sites and pathways.
Figures




Similar articles
-
p38γ activation triggers dynamical changes in allosteric docking sites.Biochemistry. 2011 Mar 1;50(8):1384-95. doi: 10.1021/bi1007518. Epub 2011 Feb 7. Biochemistry. 2011. PMID: 21235211
-
A Dynamic Switch in Inactive p38γ Leads to an Excited State on the Pathway to an Active Kinase.Biochemistry. 2019 Dec 24;58(51):5160-5172. doi: 10.1021/acs.biochem.9b00932. Epub 2019 Dec 13. Biochemistry. 2019. PMID: 31794659 Free PMC article.
-
Reciprocal allosteric regulation of p38γ and PTPN3 involves a PDZ domain-modulated complex formation.Sci Signal. 2014 Oct 14;7(347):ra98. doi: 10.1126/scisignal.2005722. Sci Signal. 2014. PMID: 25314968
-
p38γ MAPK Inflammatory and Metabolic Signaling in Physiology and Disease.Cells. 2023 Jun 21;12(13):1674. doi: 10.3390/cells12131674. Cells. 2023. PMID: 37443708 Free PMC article. Review.
-
Studying allosteric regulation in metal sensor proteins using computational methods.Adv Protein Chem Struct Biol. 2014;96:181-218. doi: 10.1016/bs.apcsb.2014.06.009. Epub 2014 Sep 6. Adv Protein Chem Struct Biol. 2014. PMID: 25443958 Review.
Cited by
-
Activation of polycystin-1 signaling by binding of stalk-derived peptide agonists.Elife. 2024 Oct 7;13:RP95992. doi: 10.7554/eLife.95992. Elife. 2024. PMID: 39373641 Free PMC article.
-
Conformational states dynamically populated by a kinase determine its function.Science. 2020 Oct 9;370(6513):eabc2754. doi: 10.1126/science.abc2754. Epub 2020 Oct 1. Science. 2020. PMID: 33004676 Free PMC article.
-
Allosteric Regulation at the Crossroads of New Technologies: Multiscale Modeling, Networks, and Machine Learning.Front Mol Biosci. 2020 Jul 9;7:136. doi: 10.3389/fmolb.2020.00136. eCollection 2020. Front Mol Biosci. 2020. PMID: 32733918 Free PMC article. Review.
-
Exploring and Learning the Universe of Protein Allostery Using Artificial Intelligence Augmented Biophysical and Computational Approaches.J Chem Inf Model. 2023 Mar 13;63(5):1413-1428. doi: 10.1021/acs.jcim.2c01634. Epub 2023 Feb 24. J Chem Inf Model. 2023. PMID: 36827465 Free PMC article. Review.
-
Assignment of Ala, Ile, LeuproS, Met, and ValproS methyl groups of the protruding domain of murine norovirus capsid protein VP1 using methyl-methyl NOEs, site directed mutagenesis, and pseudocontact shifts.Biomol NMR Assign. 2022 Apr;16(1):97-107. doi: 10.1007/s12104-022-10066-7. Epub 2022 Jan 20. Biomol NMR Assign. 2022. PMID: 35050443 Free PMC article.
References
-
- Kern D. & Zuiderweg E. R. P. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 13, 748–757 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

