Structure-function analysis and genetic interactions of the Luc7 subunit of the Saccharomyces cerevisiae U1 snRNP
- PMID: 27354704
- PMCID: PMC4986886
- DOI: 10.1261/rna.056911.116
Structure-function analysis and genetic interactions of the Luc7 subunit of the Saccharomyces cerevisiae U1 snRNP
Abstract
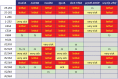
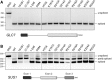
Luc7 is an essential 261-amino acid protein subunit of the Saccharomyces cerevisiae U1 snRNP. To establish structure-function relations for yeast Luc7, we conducted an in vivo mutational analysis entailing N- and C-terminal truncations and alanine scanning of phylogenetically conserved amino acids, including two putative zinc finger motifs, ZnF1 and ZnF2, and charged amino acids within the ZnF2 module. We identify Luc7-(31-246) as a minimal functional protein and demonstrate that whereas mutations of the CCHH ZnF2 motif are lethal, mutations of the ZnF1 CCCH motif and the charged residues of the ZnF2 modules are not. Though dispensable for vegetative growth in an otherwise wild-type background, the N-terminal 18-amino acid segment of Luc7 plays an important role in U1 snRNP function, evinced by our findings that its deletion (i) impaired the splicing of SUS1 pre-mRNA; (ii) was synthetically lethal absent other U1 snRNP constituents (Mud1, Nam8, the TMG cap, the C terminus of Snp1), absent the Mud2 subunit of the Msl5•Mud2 branchpoint binding complex, and when the m(7)G cap-binding site of Cbc2 was debilitated; and (iii) bypassed the need for the essential DEAD-box ATPase Prp28. Similar phenotypes were noted for ZnF1 mutations C45A, C53A, and C68A and ZnF2 domain mutations D214A, R215A, R216A, and D219A These findings highlight the contributions of the Luc7 N-terminal peptide, the ZnF1 motif, and the ZnF2 module in stabilizing the interactions of the U1 snRNP with the pre-mRNA 5' splice site and promoting the splicing of a yeast pre-mRNA, SUS1, that has a nonconsensus 5' splice site.
Keywords: Luc7; Prp28; U1 snRNP; spliceosome assembly.
© 2016 Agarwal et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Abovich N, Rosbash M. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412. - PubMed
-
- Abovich N, Liao XC, Rosbash M. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev 8: 843–854. - PubMed
-
- Chen JY, Stands L, Staley JP, Jackups RR Jr, Latus LJ, Chang TH. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell 7: 227–232. - PubMed
-
- Colot HV, Stutz F, Rosbash M. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev 10: 1699–1708. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
