Constrained Allocation Flux Balance Analysis
- PMID: 27355325
- PMCID: PMC4927118
- DOI: 10.1371/journal.pcbi.1004913
Constrained Allocation Flux Balance Analysis
Abstract
New experimental results on bacterial growth inspire a novel top-down approach to study cell metabolism, combining mass balance and proteomic constraints to extend and complement Flux Balance Analysis. We introduce here Constrained Allocation Flux Balance Analysis, CAFBA, in which the biosynthetic costs associated to growth are accounted for in an effective way through a single additional genome-wide constraint. Its roots lie in the experimentally observed pattern of proteome allocation for metabolic functions, allowing to bridge regulation and metabolism in a transparent way under the principle of growth-rate maximization. We provide a simple method to solve CAFBA efficiently and propose an "ensemble averaging" procedure to account for unknown protein costs. Applying this approach to modeling E. coli metabolism, we find that, as the growth rate increases, CAFBA solutions cross over from respiratory, growth-yield maximizing states (preferred at slow growth) to fermentative states with carbon overflow (preferred at fast growth). In addition, CAFBA allows for quantitatively accurate predictions on the rate of acetate excretion and growth yield based on only 3 parameters determined by empirical growth laws.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
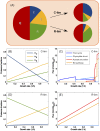
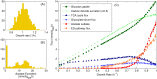
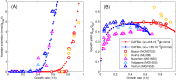



References
-
- Monod J (1949) The growth of bacterial cultures. Ann Rev Microbiol 3: 371–394 10.1146/annurev.mi.03.100149.002103 - DOI
-
- Kjeldgaard N, Kurland C (1963) The distribution of soluble and ribosomal RNA as a function of growth rate. J Mol Biol 6: 341–348 10.1016/S0022-2836(63)80093-5 - DOI
-
- Bremer H, Dennis PP (1996) Modulation of chemical composition and other parameters of the cell by growth rate. Escherichia coli and Salmonella: Cellular and Molecular Biology 2: 1553–1569
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

