HIF-1alpha Deficiency Attenuates the Cardiomyogenesis of Mouse Embryonic Stem Cells
- PMID: 27355368
- PMCID: PMC4927095
- DOI: 10.1371/journal.pone.0158358
HIF-1alpha Deficiency Attenuates the Cardiomyogenesis of Mouse Embryonic Stem Cells
Abstract
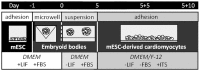
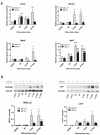
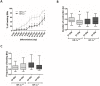
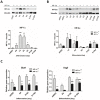
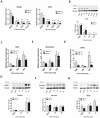
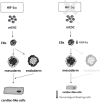
Cardiac cell formation, cardiomyogenesis, is critically dependent on oxygen availability. It is known that hypoxia, a reduced oxygen level, modulates the in vitro differentiation of pluripotent cells into cardiomyocytes via hypoxia inducible factor-1alpha (HIF-1α)-dependent mechanisms. However, the direct impact of HIF-1α deficiency on the formation and maturation of cardiac-like cells derived from mouse embryonic stem cells (mESC) in vitro remains to be elucidated. In the present study, we demonstrated that HIF-1α deficiency significantly altered the quality and quantity of mESC-derived cardiomyocytes. It was accompanied with lower mRNA and protein levels of cardiac cell specific markers (myosin heavy chains 6 and 7) and with a decreasing percentage of myosin heavy chain α and β, and cardiac troponin T-positive cells. As to structural aspects of the differentiated cardiomyocytes, the localization of contractile proteins (cardiac troponin T, myosin heavy chain α and β) and the organization of myofibrils were also different. Simultaneously, HIF-1α deficiency was associated with a lower percentage of beating embryoid bodies. Interestingly, an observed alteration in the in vitro differentiation scheme of HIF-1α deficient cells was accompanied with significantly lower expression of the endodermal marker (hepatic nuclear factor 4 alpha). These findings thus suggest that HIF-1α deficiency attenuates spontaneous cardiomyogenesis through the negative regulation of endoderm development in mESC differentiating in vitro.
Conflict of interest statement
Figures

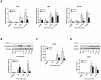
Similar articles
-
Melatonin promotes cardiomyogenesis of embryonic stem cells via inhibition of HIF-1α stabilization.J Pineal Res. 2016 Nov;61(4):493-503. doi: 10.1111/jpi.12366. Epub 2016 Oct 16. J Pineal Res. 2016. PMID: 27601067
-
Foxc1 Regulates Early Cardiomyogenesis and Functional Properties of Embryonic Stem Cell Derived Cardiomyocytes.Stem Cells. 2016 Jun;34(6):1487-500. doi: 10.1002/stem.2301. Epub 2016 Feb 18. Stem Cells. 2016. PMID: 26824887
-
Effect of oxygen on cardiac differentiation in mouse iPS cells: role of hypoxia inducible factor-1 and Wnt/beta-catenin signaling.PLoS One. 2013 Nov 12;8(11):e80280. doi: 10.1371/journal.pone.0080280. eCollection 2013. PLoS One. 2013. PMID: 24265804 Free PMC article.
-
Exogenous expression of HIF-1 alpha promotes cardiac differentiation of embryonic stem cells.J Mol Cell Cardiol. 2010 Jun;48(6):1129-37. doi: 10.1016/j.yjmcc.2010.01.015. Epub 2010 Jan 29. J Mol Cell Cardiol. 2010. PMID: 20116384
-
Role of hypoxia inducible factor-1α for interferon synthesis in mouse dendritic cells.Biol Chem. 2013 Apr;394(4):495-505. doi: 10.1515/hsz-2012-0320. Biol Chem. 2013. PMID: 23362200 Review.
Cited by
-
Conductive Polymer PEDOT:PSS-Based Platform for Embryonic Stem-Cell Differentiation.Int J Mol Sci. 2022 Jan 20;23(3):1107. doi: 10.3390/ijms23031107. Int J Mol Sci. 2022. PMID: 35163031 Free PMC article.
-
Use of Therapeutic RNAs to Accelerate Wound Healing in Diabetic Rabbit Wounds.Adv Wound Care (New Rochelle). 2024 Sep;13(9):435-445. doi: 10.1089/wound.2023.0056. Epub 2024 Mar 1. Adv Wound Care (New Rochelle). 2024. PMID: 38183631
-
Hypoxia preconditioning promotes cardiac stem cell survival and cardiogenic differentiation in vitro involving activation of the HIF-1α/apelin/APJ axis.Stem Cell Res Ther. 2017 Sep 29;8(1):215. doi: 10.1186/s13287-017-0673-4. Stem Cell Res Ther. 2017. PMID: 28962638 Free PMC article.
-
Recent advances and challenges on application of tissue engineering for treatment of congenital heart disease.PeerJ. 2018 Oct 25;6:e5805. doi: 10.7717/peerj.5805. eCollection 2018. PeerJ. 2018. PMID: 30386701 Free PMC article.
-
HIF-1α in Osteoarthritis: From Pathogenesis to Therapeutic Implications.Front Pharmacol. 2022 Jul 5;13:927126. doi: 10.3389/fphar.2022.927126. eCollection 2022. Front Pharmacol. 2022. PMID: 35865944 Free PMC article. Review.
References
-
- Braunwald E, Bristow MR. Congestive heart failure: fifty years of progress. Circulation. 2000;102(20 Suppl 4):IV14–23. Epub 2000/11/18. . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

