Structural Analysis using SHALiPE to Reveal RNA G-Quadruplex Formation in Human Precursor MicroRNA
- PMID: 27355429
- PMCID: PMC6680278
- DOI: 10.1002/anie.201603562
Structural Analysis using SHALiPE to Reveal RNA G-Quadruplex Formation in Human Precursor MicroRNA
Abstract
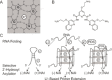
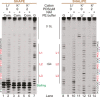
RNA G-quadruplex (rG4) structures are of fundamental importance to biology. A novel approach is introduced to detect and structurally map rG4s at single-nucleotide resolution in RNAs. The approach, denoted SHALiPE, couples selective 2'-hydroxyl acylation with lithium ion-based primer extension, and identifies characteristic structural fingerprints for rG4 mapping. We apply SHALiPE to interrogate the human precursor microRNA 149, and reveal the formation of an rG4 structure in this non-coding RNA. Additional analyses support the SHALiPE results and uncover that this rG4 has a parallel topology, is thermally stable, and is conserved in mammals. An in vitro Dicer assay shows that this rG4 inhibits Dicer processing, supporting the potential role of rG4 structures in microRNA maturation and post-transcriptional regulation of mRNAs.
Keywords: Dicer processing; G-quadruplexes; RNA structure; precursor miRNA; structure probing.
© 2016 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

