TCGA: Increased oncoprotein coding region mutations correlate with a greater expression of apoptosis-effector genes and a positive outcome for stomach adenocarcinoma
- PMID: 27355872
- PMCID: PMC4993568
- DOI: 10.1080/15384101.2016.1195532
TCGA: Increased oncoprotein coding region mutations correlate with a greater expression of apoptosis-effector genes and a positive outcome for stomach adenocarcinoma
Abstract
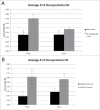
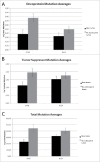
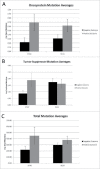
Oncogene mutations are primarily thought to facilitate uncontrolled cell growth. However, overexpression of oncoproteins likely leads to apoptosis in a feed forward mechanism, whereby a certain level of oncoprotein leads to the activation of pro-proliferation effector genes and higher levels lead to activation of pro-apoptotic effector genes. TCGA STAD barcodes having no oncoprotein coding region mutations represented reduced expression of the apoptosis-effector genes compared with barcodes with multiple oncoprotein coding region mutations. Furthermore, STAD barcodes in a "no-subsequent tumor" group, representing 224 samples, and in a "positive outcome" group, had more oncoprotein coding regions mutated, on average, than barcodes of the new tumor and negative outcome groups, respectively. BRAF, CTNNB1, KRAS and MTOR coding region mutations (as a group) had the strongest association with the no-subsequent tumor group. Tumor suppressor coding region mutations were also correlated with no-subsequent tumor. These results are consistent with an oncoprotein-mediated, feed-forward mechanism of apoptosis in patients. Importantly, the no-subsequent tumor group also had more overall mutations. This result leads to considerations of unhealthy cells or cells with more neo-antigens for immune rejection. However, a probabilistic aspect of mutagenesis is also consistent with more oncoprotein and tumor suppressor protein mutations, in cases of more overall mutations, and thus a higher likelihood of activation of feed forward apoptosis pathways.
Keywords: apoptosis-effector genes; feed-forward apoptosis; oncoprotein mediated apoptosis; stomach adenocarcinoma; tumor suppressor proteins.
Figures
Similar articles
-
Identification of aberrantly expressed long non-coding RNAs in stomach adenocarcinoma.Oncotarget. 2017 Jul 25;8(30):49201-49216. doi: 10.18632/oncotarget.17329. Oncotarget. 2017. PMID: 28484081 Free PMC article.
-
Inactivating mutation of the pro-apoptotic gene BID in gastric cancer.J Pathol. 2004 Apr;202(4):439-45. doi: 10.1002/path.1532. J Pathol. 2004. PMID: 15095271
-
Hypermethylation of XIAP-associated factor 1, a putative tumor suppressor gene from the 17p13.2 locus, in human gastric adenocarcinomas.Cancer Res. 2003 Nov 1;63(21):7068-75. Cancer Res. 2003. PMID: 14612497
-
[Peculiarities of regulation of tumour cell apoptosis].Vestn Ross Akad Med Nauk. 2008;(10):15-20. Vestn Ross Akad Med Nauk. 2008. PMID: 19140394 Review. Russian.
-
Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death.IUBMB Life. 2010 Dec;62(12):896-905. doi: 10.1002/iub.396. IUBMB Life. 2010. PMID: 21154818 Review.
Cited by
-
Retinoblastoma cells activate the AKT pathway and are vulnerable to the PI3K/mTOR inhibitor NVP-BEZ235.Oncotarget. 2017 Jun 13;8(24):38084-38098. doi: 10.18632/oncotarget.16970. Oncotarget. 2017. PMID: 28445155 Free PMC article.
-
Retinoblastoma tumor cell proliferation is negatively associated with an immune gene expression signature and increased immune cells.Lab Invest. 2021 Jun;101(6):701-718. doi: 10.1038/s41374-021-00573-x. Epub 2021 Mar 3. Lab Invest. 2021. PMID: 33658609
-
Identification of specific feed-forward apoptosis mechanisms and associated higher survival rates for low grade glioma and lung squamous cell carcinoma.J Cancer Res Clin Oncol. 2018 Mar;144(3):459-468. doi: 10.1007/s00432-017-2569-1. Epub 2018 Jan 5. J Cancer Res Clin Oncol. 2018. PMID: 29305708 Free PMC article.
-
Elucidating feed-forward apoptosis signatures in breast cancer datasets: Higher FOS expression associated with a better outcome.Oncol Lett. 2018 Aug;16(2):2757-2763. doi: 10.3892/ol.2018.8957. Epub 2018 Jun 12. Oncol Lett. 2018. PMID: 30013671 Free PMC article.
-
An age-based, RNA expression paradigm for survival biomarker identification for pediatric neuroblastoma and acute lymphoblastic leukemia.Cancer Cell Int. 2019 Mar 27;19:73. doi: 10.1186/s12935-019-0790-5. eCollection 2019. Cancer Cell Int. 2019. PMID: 30962767 Free PMC article.
References
-
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 1996; 85(4):549-61; PMID:8653790; http://dx.doi.org/10.1016/S0092-8674(00)81255-6 - DOI - PubMed
-
- Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 1996; 85(4):537-48; PMID:8653789; http://dx.doi.org/10.1016/S0092-8674(00)81254-4 - DOI - PubMed
-
- Oswald F, Dobner T, Lipp M. The E2F transcription factor activates a replication-dependent human H2A gene in early S phase of the cell cycle. Mol Cell Biol 1996; 16(5):1889-95; PMID:8628255; http://dx.doi.org/10.1128/MCB.16.5.1889 - DOI - PMC - PubMed
-
- Banerjee D, Ercikan-Abali E, Waltham M, Schnieders B, Hochhauser D, Li WW, Fan J, Gorlick R, Goker E, Bertino JR. Molecular mechanisms of resistance to antifolates, a review. Acta Biochimica Polonica 1995; 42(4):457-64; PMID:8852336 - PubMed
-
- Kirch HC, Putzer B, Schwabe G, Gnauck HK, Schulte Holthausen H. Regulation of adenovirus 12 E1A transcription: E2F and ATF motifs in the E1A promoter bind nuclear protein complexes including E2F1, DP-1, cyclin A and/or RB and mediate transcriptional (auto)activation. Cell Mol Biol Res 1993; 39(8):705-16; PMID:7951410 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
