Transcriptome comparison reveals a genetic network regulating the lower temperature limit in fish
- PMID: 27356472
- PMCID: PMC4928090
- DOI: 10.1038/srep28952
Transcriptome comparison reveals a genetic network regulating the lower temperature limit in fish
Abstract
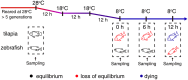
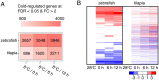
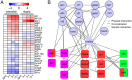
Transcriptional plasticity is a major driver of phenotypic differences between species. The lower temperature limit (LTL), namely the lower end of survival temperature, is an important trait delimiting the geographical distribution of a species, however, the genetic mechanisms are poorly understood. We investigated the inter-species transcriptional diversification in cold responses between zebrafish Danio rerio and tilapia Oreochromis niloticus, which were reared at a common temperature (28 °C) but have distinct LTLs. We identified significant expressional divergence between the two species in the orthologous genes from gills when the temperature cooled to the LTL of tilapia (8 °C). Five KEGG pathways were found sequentially over-represented in the zebrafish/tilapia divergently expressed genes in the duration (12 hour) of 8 °C exposure, forming a signaling cascade from metabolic regulation to apoptosis via FoxO signaling. Consistently, we found differential progression of apoptosis in the gills of the two species in which zebrafish manifested a delayed and milder apoptotic phenotype than tilapia, corresponding with a lower LTL of zebrafish. We identified diverged expression in 25 apoptosis-related transcription factors between the two species which forms an interacting network with diverged factors involving the FoxO signaling and metabolic regulation. We propose a genetic network which regulates LTL in fishes.
Figures



References
-
- Carroll S. B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 (2008). - PubMed
-
- Hoekstra H. E. & Coyne J. A. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995–1016 (2007). - PubMed
-
- Cossins A. R. & Crawford D. L. Fish as models for environmental genomics. Nat. Rev. Genet. 6, 324–333 (2005). - PubMed
-
- Donaldson M. R., Cooke S. J., Patterson D. A. & Macdonald J. S. Cold shock and fish. J. Fish Biol. 73, 1491–1530, 10.1111/j.1095-8649.2008.02061.x (2008). - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

