Negative selection maintains transcription factor binding motifs in human cancer
- PMID: 27356864
- PMCID: PMC4928157
- DOI: 10.1186/s12864-016-2728-9
Negative selection maintains transcription factor binding motifs in human cancer
Abstract
Background: Somatic mutations in cancer cells affect various genomic elements disrupting important cell functions. In particular, mutations in DNA binding sites recognized by transcription factors can alter regulator binding affinities and, consequently, expression of target genes. A number of promoter mutations have been linked with an increased risk of cancer. Cancer somatic mutations in binding sites of selected transcription factors have been found under positive selection. However, action and significance of negative selection in non-coding regions remain controversial.
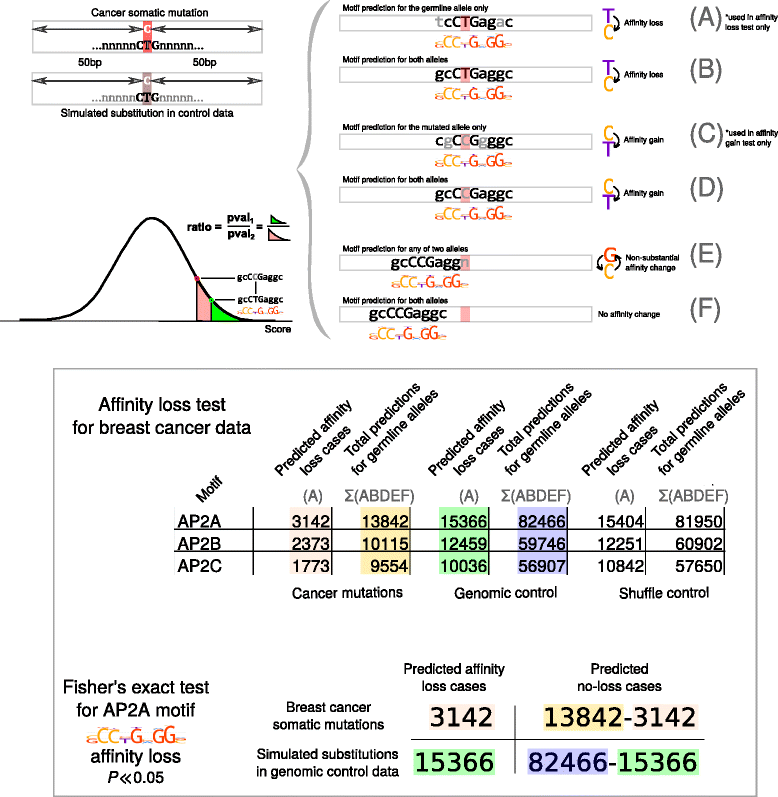
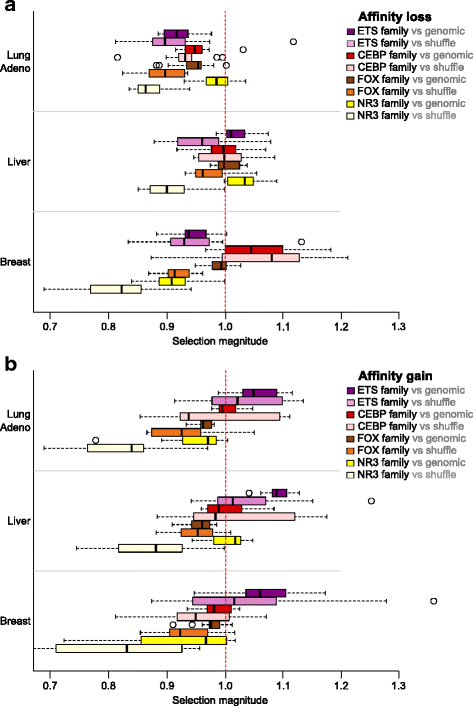
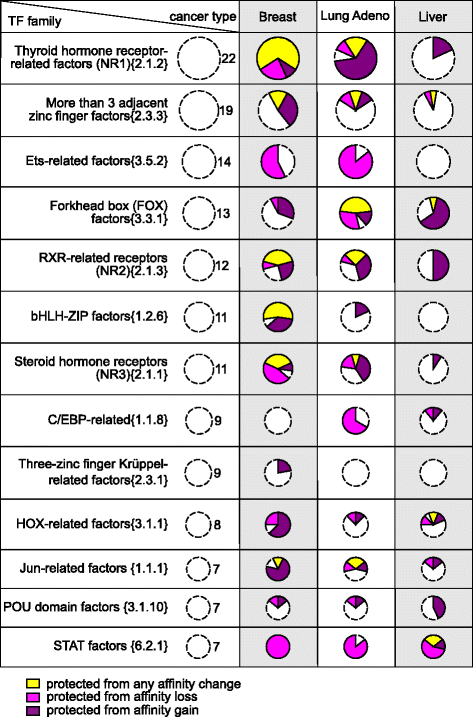
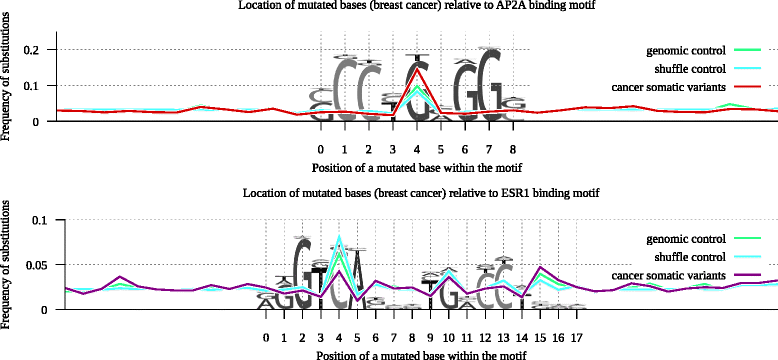
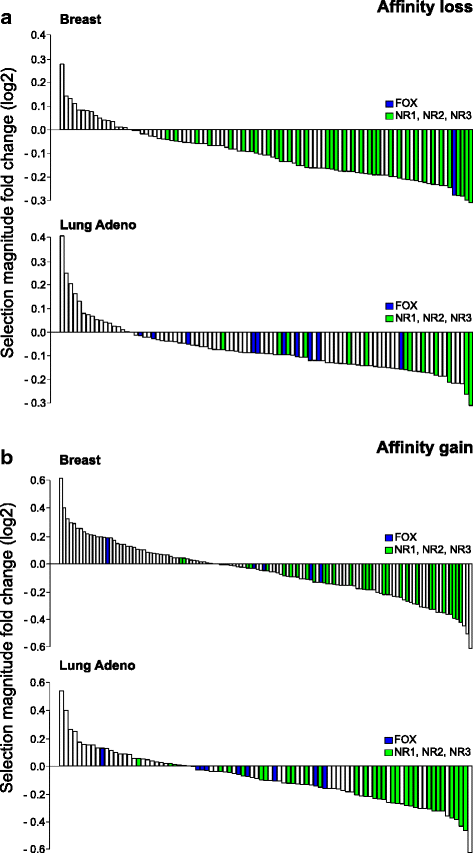
Results: Here we present analysis of transcription factor binding motifs co-localized with non-coding variants. To avoid statistical bias we account for mutation signatures of different cancer types. For many transcription factors, including multiple members of FOX, HOX, and NR families, we show that human cancers accumulate fewer mutations than expected by chance that increase or decrease affinity of predicted binding sites. Such stability of binding motifs is even more exhibited in DNase accessible regions.
Conclusions: Our data demonstrate negative selection against binding sites alterations and suggest that such selection pressure protects cancer cells from rewiring of regulatory circuits. Further analysis of transcription factors with conserved binding motifs can reveal cell regulatory pathways crucial for the survivability of various human cancers.
Keywords: Cancer somatic mutations; DNA motifs; Negative selection; Transcription factor binding sites.
Figures

References
-
- Bell RJA, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, Wiencke JK, Wrensch MR, Chang SM, Walsh KM, Myong S, Song JS, Costello JF. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. - DOI - PMC - PubMed
-
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He T-C, He Y, Hruban RH, Jallo GI, Mandahl N, Meeker AK, Mertens F, Netto GJ, Rasheed BA, Riggins GJ, Rosenquist TA, Schiffman M, Shih I-M, Theodorescu D, Torbenson MS, Velculescu VE, Wang T-L, Wentzensen N, Wood LD, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

