Borrowed alleles and convergence in serpentine adaptation
- PMID: 27357660
- PMCID: PMC4961121
- DOI: 10.1073/pnas.1600405113
Borrowed alleles and convergence in serpentine adaptation
Abstract
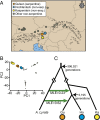
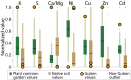
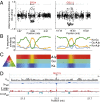
Serpentine barrens represent extreme hazards for plant colonists. These sites are characterized by high porosity leading to drought, lack of essential mineral nutrients, and phytotoxic levels of metals. Nevertheless, nature forged populations adapted to these challenges. Here, we use a population-based evolutionary genomic approach coupled with elemental profiling to assess how autotetraploid Arabidopsis arenosa adapted to a multichallenge serpentine habitat in the Austrian Alps. We first demonstrate that serpentine-adapted plants exhibit dramatically altered elemental accumulation levels in common conditions, and then resequence 24 autotetraploid individuals from three populations to perform a genome scan. We find evidence for highly localized selective sweeps that point to a polygenic, multitrait basis for serpentine adaptation. Comparing our results to a previous study of independent serpentine colonizations in the closely related diploid Arabidopsis lyrata in the United Kingdom and United States, we find the highest levels of differentiation in 11 of the same loci, providing candidate alleles for mediating convergent evolution. This overlap between independent colonizations in different species suggests that a limited number of evolutionary strategies are suited to overcome the multiple challenges of serpentine adaptation. Interestingly, we detect footprints of selection in A. arenosa in the context of substantial gene flow from nearby off-serpentine populations of A. arenosa, as well as from A. lyrata In several cases, quantitative tests of introgression indicate that some alleles exhibiting strong selective sweep signatures appear to have been introgressed from A. lyrata This finding suggests that migrant alleles may have facilitated adaptation of A. arenosa to this multihazard environment.
Keywords: adaptation; gene flow; plant; population genomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Proctor J, Woodell SR. The ecology of serpentine soils. Adv Ecol Res. 1975;9:255–366.
-
- Brady KU, Kruckeberg AR, Bradshaw HD., Jr Evolutionary ecology of plant adaptation to serpentine soils. Annu Rev Ecol Evol Syst. 2005;36(1):243–266.
-
- Harrison S, Rajakaruna N. Serpentine: The Evolution and Ecology of a Model System. Univ of California Press; Berkeley: 2011.
-
- Vlamis J, Jenny H. Calcium deficiency in serpentine soils as revealed by adsorbent technique. Science. 1948;107(2786):549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

