Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer
- PMID: 27357679
- PMCID: PMC4961202
- DOI: 10.1073/pnas.1601571113
Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer
Abstract
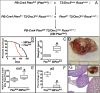
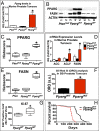
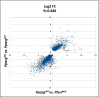
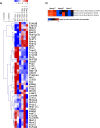
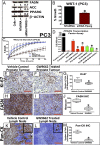
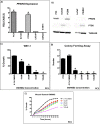
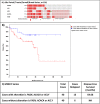
Prostate cancer (CaP) is the most common adult male cancer in the developed world. The paucity of biomarkers to predict prostate tumor biology makes it important to identify key pathways that confer poor prognosis and guide potential targeted therapy. Using a murine forward mutagenesis screen in a Pten-null background, we identified peroxisome proliferator-activated receptor gamma (Pparg), encoding a ligand-activated transcription factor, as a promoter of metastatic CaP through activation of lipid signaling pathways, including up-regulation of lipid synthesis enzymes [fatty acid synthase (FASN), acetyl-CoA carboxylase (ACC), ATP citrate lyase (ACLY)]. Importantly, inhibition of PPARG suppressed tumor growth in vivo, with down-regulation of the lipid synthesis program. We show that elevated levels of PPARG strongly correlate with elevation of FASN in human CaP and that high levels of PPARG/FASN and PI3K/pAKT pathway activation confer a poor prognosis. These data suggest that CaP patients could be stratified in terms of PPARG/FASN and PTEN levels to identify patients with aggressive CaP who may respond favorably to PPARG/FASN inhibition.
Keywords: PPARG; PTEN; Sleeping Beauty; metastasis; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

