Digital Quantification of DNA Replication and Chromosome Segregation Enables Determination of Antimicrobial Susceptibility after only 15 Minutes of Antibiotic Exposure
- PMID: 27357747
- PMCID: PMC5215780
- DOI: 10.1002/anie.201602763
Digital Quantification of DNA Replication and Chromosome Segregation Enables Determination of Antimicrobial Susceptibility after only 15 Minutes of Antibiotic Exposure
Erratum in
-
Corrigendum: Digital Quantification of DNA Replication and Chromosome Segregation Enables Determination of Antimicrobial Susceptibility after only 15 Minutes of Antibiotic Exposure.Angew Chem Int Ed Engl. 2017 Sep 18;56(39):11675. doi: 10.1002/anie.201707742. Angew Chem Int Ed Engl. 2017. PMID: 28914476 No abstract available.
Abstract
Rapid antimicrobial susceptibility testing (AST) would decrease misuse and overuse of antibiotics. The "holy grail" of AST is a phenotype-based test that can be performed within a doctor visit. Such a test requires the ability to determine a pathogen's susceptibility after only a short antibiotic exposure. Herein, digital PCR (dPCR) was employed to test whether measuring DNA replication of the target pathogen through digital single-molecule counting would shorten the required time of antibiotic exposure. Partitioning bacterial chromosomal DNA into many small volumes during dPCR enabled AST results after short exposure times by 1) precise quantification and 2) a measurement of how antibiotics affect the states of macromolecular assembly of bacterial chromosomes. This digital AST (dAST) determined susceptibility of clinical isolates from urinary tract infections (UTIs) after 15 min of exposure for all four antibiotic classes relevant to UTIs. This work lays the foundation to develop a rapid, point-of-care AST and strengthen global antibiotic stewardship.
Keywords: DNA replication; analytical methods; antibiotics; polymerase chain reaction; susceptibility.
© 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

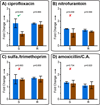


References
-
- CDC. Antibiotic Resistance Threats in the United States. U. S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2013. p. 114.
- Laxminarayan R, et al. Lancet Infect. Dis. 2013;13:1057–1098. - PubMed
-
- White House. National Action Plan for Combating Antibiotic-Resistant Bacteria. Washington D.C.: 2015. p. 62.
-
- Roberts RR, et al. Clin. Infect. Dis. 2009;49:1175–1184. - PubMed
-
- Besant JD, Sargent EH, Kelley SO. Lab Chip. 2015;15:2799–2807. - PubMed
- Lu Y, Gao J, Zhang DD, Gau V, Liao JC, Wong PK. Anal. Chem. 2013;85:3971–3976. - PMC - PubMed
- Zhu C, Yang Q, Liu L, Wang S. Angew. Chem. Int. Ed. Engl. 2011;50:9607–9610. - PubMed
- Waldeisen JR, Wang T, Mitra D, Lee LP. PLoS One. 2011;6:e28528. - PMC - PubMed
- Rolain JM, Mallet MN, Fournier PE, Raoult D. J. Antimicrob. Chemother. 2004;54:538–541. - PubMed
- Mezger A, Gullberg E, Goransson J, Zorzet A, Herthnek D, Tano E, Nilsson M, Andersson DI. J. Clin. Microbiol. 2015;53:425–432. - PMC - PubMed
- Mach KE, Mohan R, Baron EJ, Shih MC, Gau V, Wong PK, Liao JC. J. Urol. 2011;185:148–153. - PMC - PubMed
- Ivančić V, et al. J. Clin. Microbiol. 2008;46:1213–1219. - PMC - PubMed
- Halford C, Gonzalez R, Campuzano S, Hu B, Babbitt JT, Liu J, Wang J, Churchill BM, Haake DA. Antimicrob. Agents Chemother. 2013;57:936–943. - PMC - PubMed
- Barczak AK, et al. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6217–6222. - PMC - PubMed
- Sinn I, Kinnunen P, Albertson T, McNaughton BH, Newton DW, Burns MA, Kopelman R. Lab Chip. 2011;11:2604–2611. - PubMed
- Mann TS, Mikkelsen SR. Anal. Chem. 2008;80:843–848. - PubMed
- Tang Y, Zhen L, Liu J, Wu J. Anal. Chem. 2013;85:2787–2794. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

