An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling
- PMID: 27357749
- PMCID: PMC4928087
- DOI: 10.1038/srep28941
An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling
Erratum in
-
Author Correction: An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signalling.Sci Rep. 2026 Jan 8;16(1):874. doi: 10.1038/s41598-025-33609-z. Sci Rep. 2026. PMID: 41507252 Free PMC article. No abstract available.
Abstract
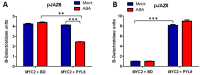
Abscisic acid (ABA) is a plant hormone that mediates abiotic stress tolerance and regulates growth and development. ABA binds to members of the PYL/RCAR ABA receptor family that initiate signal transduction inhibiting type 2C protein phosphatases. Although crosstalk between ABA and the hormone Jasmonic Acid (JA) has been shown, the molecular entities that mediate this interaction have yet to be fully elucidated. We report a link between ABA and JA signaling through a direct interaction of the ABA receptor PYL6 (RCAR9) with the basic helix-loop-helix transcription factor MYC2. PYL6 and MYC2 interact in yeast two hybrid assays and the interaction is enhanced in the presence of ABA. PYL6 and MYC2 interact in planta based on bimolecular fluorescence complementation and co-immunoprecipitation of the proteins. Furthermore, PYL6 was able to modify transcription driven by MYC2 using JAZ6 and JAZ8 DNA promoter elements in yeast one hybrid assays. Finally, pyl6 T-DNA mutant plants show an increased sensitivity to the addition of JA along with ABA in cotyledon expansion experiments. Overall, the present study identifies a direct mechanism for transcriptional modulation mediated by an ABA receptor different from the core ABA signaling pathway, and a putative mechanistic link connecting ABA and JA signaling pathways.
Figures




References
-
- Lopez-Molina L., Mongrand S. & Chua N. H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 98, 4782–4787, doi: 10.1073/pnas.081594298 (2001). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

