Proteomic analysis of breast tumors confirms the mRNA intrinsic molecular subtypes using different classifiers: a large-scale analysis of fresh frozen tissue samples
- PMID: 27357824
- PMCID: PMC4928264
- DOI: 10.1186/s13058-016-0732-2
Proteomic analysis of breast tumors confirms the mRNA intrinsic molecular subtypes using different classifiers: a large-scale analysis of fresh frozen tissue samples
Abstract
Background: Breast cancer is a complex and heterogeneous disease that is usually characterized by histological parameters such as tumor size, cellular arrangements/rearrangments, necrosis, nuclear grade and the mitotic index, leading to a set of around twenty subtypes. Together with clinical markers such as hormone receptor status, this classification has considerable prognostic value but there is a large variation in patient response to therapy. Gene expression profiling has provided molecular profiles characteristic of distinct subtypes of breast cancer that reflect the divergent cellular origins and degree of progression.
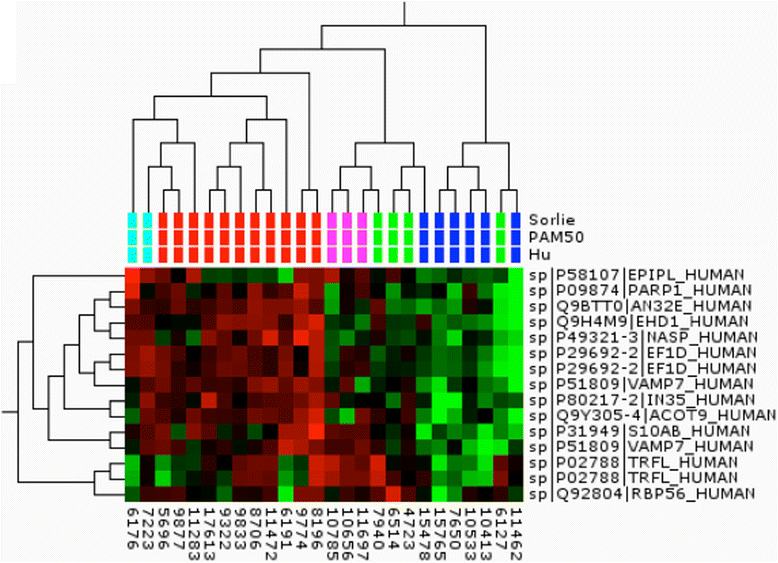
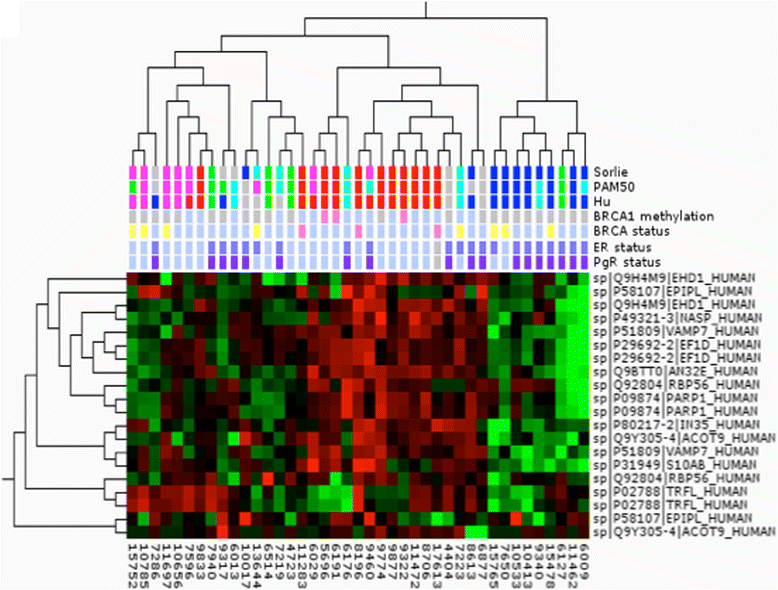
Methods: Here we present a large-scale proteomic and transcriptomic profiling study of 477 sporadic and hereditary breast cancer tumors with matching mRNA expression analysis. Unsupervised hierarchal clustering was performed and selected proteins from large-scale tandem mass spectrometry (MS/MS) analysis were transferred into a highly multiplexed targeted selected reaction monitoring assay to classify tumors using a hierarchal cluster and support vector machine with leave one out cross-validation.
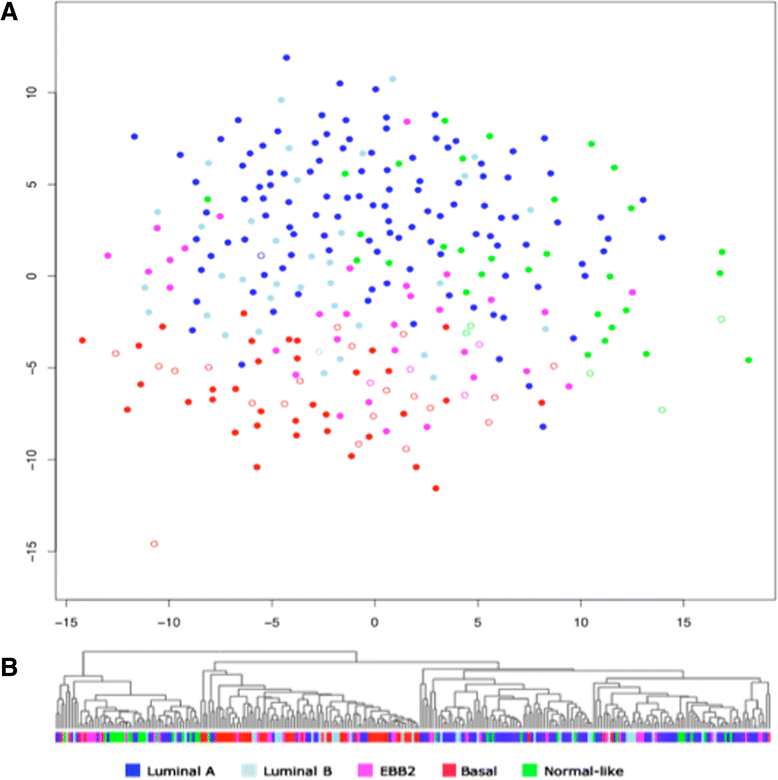
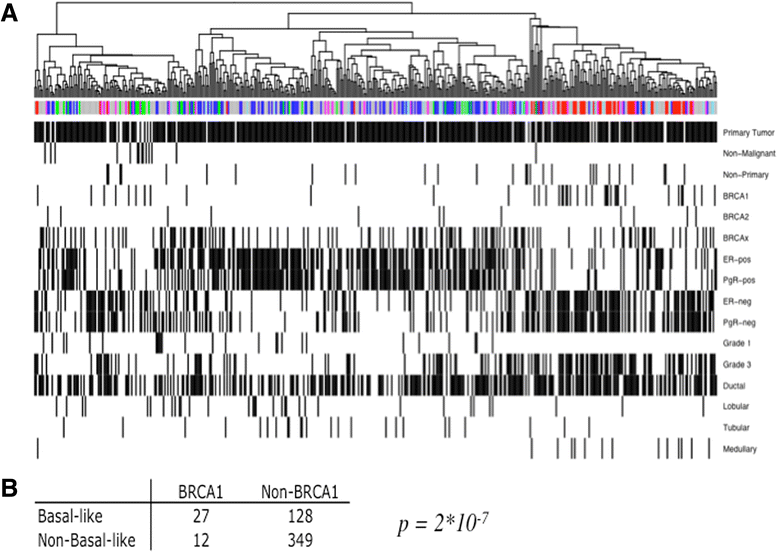
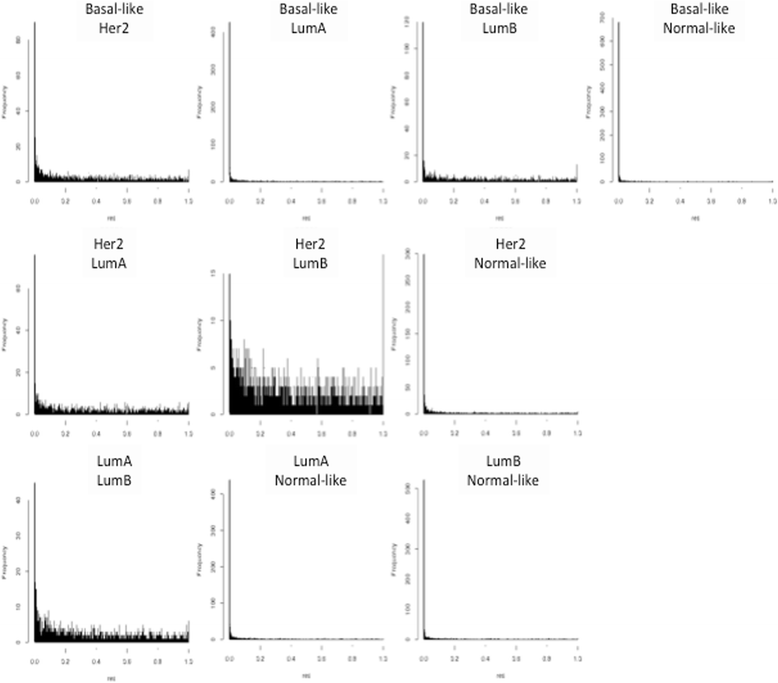
Results: The subgroups formed upon unsupervised clustering agree very well with groups found at transcriptional level; however, the classifiers (genes or their respective protein products) differ almost entirely between the two datasets. In-depth analysis shows clear differences in pathways unique to each type, which may lie behind their different clinical outcomes. Targeted mass spectrometry analysis and supervised clustering correlate very well with subgroups determined by RNA classification and show convincing agreement with clinical parameters.
Conclusions: This work demonstrates the merits of protein expression profiling for breast cancer stratification. These findings have important implications for the use of genomics and expression analysis for the prediction of protein expression, such as receptor status and drug target expression. The highly multiplexed MS assay is easily implemented in standard clinical chemistry practice, allowing rapid and cheap characterization of tumor tissue suitable for directing the choice of treatment.
Keywords: Breast cancer; Mass spectrometry; Molecular subtyping; Proteomics; Transcriptomics.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

