KinMutRF: a random forest classifier of sequence variants in the human protein kinase superfamily
- PMID: 27357839
- PMCID: PMC4928150
- DOI: 10.1186/s12864-016-2723-1
KinMutRF: a random forest classifier of sequence variants in the human protein kinase superfamily
Abstract
Background: The association between aberrant signal processing by protein kinases and human diseases such as cancer was established long time ago. However, understanding the link between sequence variants in the protein kinase superfamily and the mechanistic complex traits at the molecular level remains challenging: cells tolerate most genomic alterations and only a minor fraction disrupt molecular function sufficiently and drive disease.
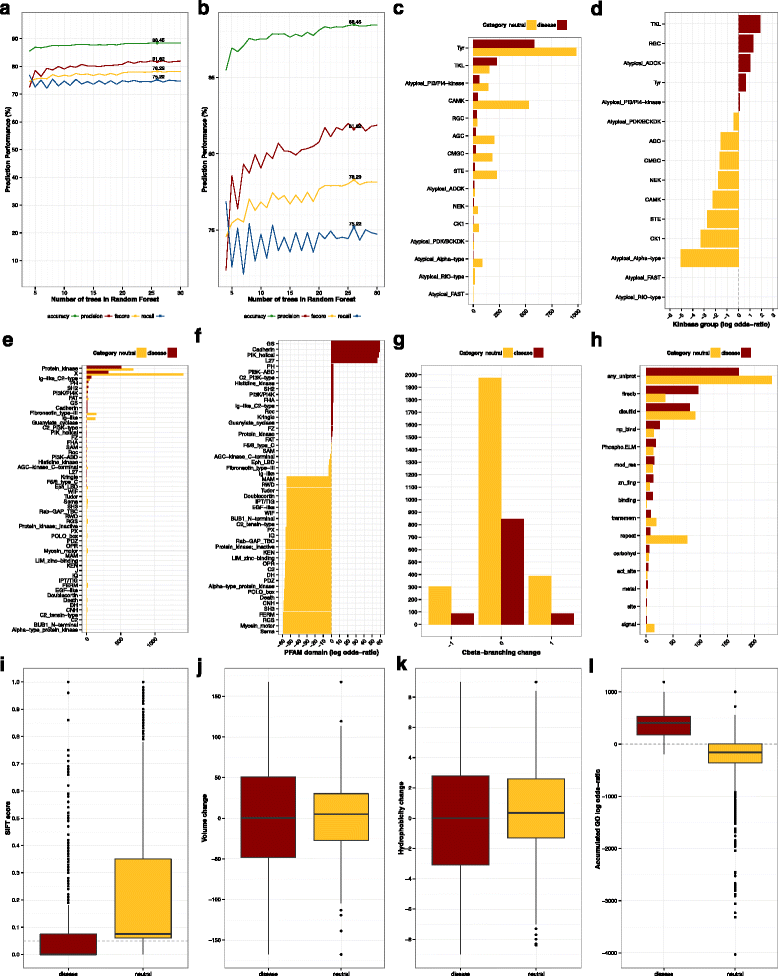
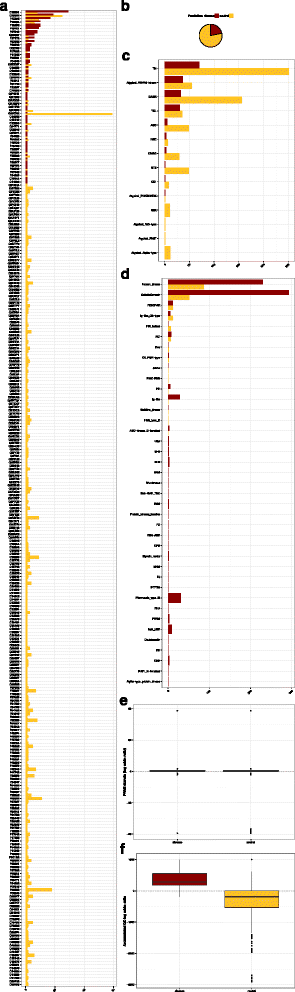
Results: KinMutRF is a novel random-forest method to automatically identify pathogenic variants in human kinases. Twenty six decision trees implemented as a random forest ponder a battery of features that characterize the variants: a) at the gene level, including membership to a Kinbase group and Gene Ontology terms; b) at the PFAM domain level; and c) at the residue level, the types of amino acids involved, changes in biochemical properties, functional annotations from UniProt, Phospho.ELM and FireDB. KinMutRF identifies disease-associated variants satisfactorily (Acc: 0.88, Prec:0.82, Rec:0.75, F-score:0.78, MCC:0.68) when trained and cross-validated with the 3689 human kinase variants from UniProt that have been annotated as neutral or pathogenic. All unclassified variants were excluded from the training set. Furthermore, KinMutRF is discussed with respect to two independent kinase-specific sets of mutations no included in the training and testing, Kin-Driver (643 variants) and Pon-BTK (1495 variants). Moreover, we provide predictions for the 848 protein kinase variants in UniProt that remained unclassified. A public implementation of KinMutRF, including documentation and examples, is available online ( http://kinmut2.bioinfo.cnio.es ). The source code for local installation is released under a GPL version 3 license, and can be downloaded from https://github.com/Rbbt-Workflows/KinMut2 .
Conclusions: KinMutRF is capable of classifying kinase variation with good performance. Predictions by KinMutRF compare favorably in a benchmark with other state-of-the-art methods (i.e. SIFT, Polyphen-2, MutationAssesor, MutationTaster, LRT, CADD, FATHMM, and VEST). Kinase-specific features rank as the most elucidatory in terms of information gain and are likely the improvement in prediction performance. This advocates for the development of family-specific classifiers able to exploit the discriminatory power of features unique to individual protein families.
Keywords: Functional impact; Pathogenicity prediction; Protein kinases; Variant prioritization; X-linked agammaglobulinemia.
Figures
Similar articles
-
wKinMut-2: Identification and Interpretation of Pathogenic Variants in Human Protein Kinases.Hum Mutat. 2016 Jan;37(1):36-42. doi: 10.1002/humu.22914. Epub 2015 Oct 20. Hum Mutat. 2016. PMID: 26443060
-
Prioritization of pathogenic mutations in the protein kinase superfamily.BMC Genomics. 2012 Jun 18;13 Suppl 4(Suppl 4):S3. doi: 10.1186/1471-2164-13-S4-S3. BMC Genomics. 2012. PMID: 22759651 Free PMC article.
-
wKinMut: an integrated tool for the analysis and interpretation of mutations in human protein kinases.BMC Bioinformatics. 2013 Nov 29;14:345. doi: 10.1186/1471-2105-14-345. BMC Bioinformatics. 2013. PMID: 24289158 Free PMC article.
-
Statistical geometry based prediction of nonsynonymous SNP functional effects using random forest and neuro-fuzzy classifiers.Proteins. 2008 Jun;71(4):1930-9. doi: 10.1002/prot.21838. Proteins. 2008. PMID: 18186470
-
Molecular genetic epidemiology of human diseases: from patterns to predictions.Hum Genet. 2014 Apr;133(4):425-30. doi: 10.1007/s00439-013-1396-y. Epub 2013 Nov 19. Hum Genet. 2014. PMID: 24241280 Review.
Cited by
-
VariBench, new variation benchmark categories and data sets.Front Bioinform. 2023 Sep 19;3:1248732. doi: 10.3389/fbinf.2023.1248732. eCollection 2023. Front Bioinform. 2023. PMID: 37795169 Free PMC article. No abstract available.
-
Analysis of somatic mutations across the kinome reveals loss-of-function mutations in multiple cancer types.Sci Rep. 2017 Jul 25;7(1):6418. doi: 10.1038/s41598-017-06366-x. Sci Rep. 2017. PMID: 28743916 Free PMC article.
-
Gain-of-Function Variomics and Multi-omics Network Biology for Precision Medicine.Methods Mol Biol. 2023;2660:357-372. doi: 10.1007/978-1-0716-3163-8_24. Methods Mol Biol. 2023. PMID: 37191809 Free PMC article.
-
IDRMutPred: predicting disease-associated germline nonsynonymous single nucleotide variants (nsSNVs) in intrinsically disordered regions.Bioinformatics. 2020 Dec 22;36(20):4977-4983. doi: 10.1093/bioinformatics/btaa618. Bioinformatics. 2020. PMID: 32756939 Free PMC article.
-
VarI-SIG 2015: methods for personalized medicine - the role of variant interpretation in research and diagnostics.BMC Genomics. 2016 Jun 23;17 Suppl 2(Suppl 2):425. doi: 10.1186/s12864-016-2721-3. BMC Genomics. 2016. PMID: 27357578 Free PMC article. No abstract available.
References
-
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. - DOI - PMC - PubMed
-
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JKV, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. - DOI - PubMed
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JKV, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PVK, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. - DOI - PubMed
-
- Creixell P, Schoof EM, Simpson CD, Longden J, Miller CJ, Lou HJ, Perryman L, Cox TR, Zivanovic N, Palmeri A, Wesolowska-Andersen A, Helmer-Citterich M, Ferkinghoff-Borg J, Itamochi H, Bodenmiller B, Erler JT, Turk BE, Linding R. Kinome-wide Decoding of Network-Attacking Mutations Rewiring Cancer Signaling. Cell. 2015;163:202–217. doi: 10.1016/j.cell.2015.08.056. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

