Effect of low doses of estradiol and tamoxifen on breast cancer cell karyotypes
- PMID: 27357940
- PMCID: PMC5064758
- DOI: 10.1530/ERC-16-0078
Effect of low doses of estradiol and tamoxifen on breast cancer cell karyotypes
Abstract
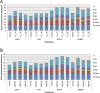
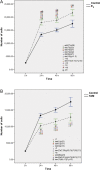
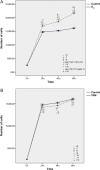
Evidence supports a role of 17&-estradiol (E2) in carcinogenesis and the large majority of breast carcinomas are dependent on estrogen. The anti-estrogen tamoxifen (TAM) is widely used for both treatment and prevention of breast cancer; however, it is also carcinogenic in human uterus and rat liver, highlighting the profound complexity of its actions. The nature of E2- or TAM-induced chromosomal damage has been explored using relatively high concentrations of these agents, and only some numerical aberrations and chromosomal breaks have been analyzed. This study aimed to determine the effects of low doses of E2 and TAM (10(&8 )mol L(&1) and 10(&6 )mol L(&1) respectively) on karyotypes of MCF7, T47D, BT474, and SKBR3 breast cancer cells by comparing the results of conventional karyotyping and multi-FISH painting with cell proliferation. Estrogen receptor (ER)-positive (+) cells showed an increase in cell proliferation after E2 treatment (MCF7, T47D, and BT474) and a decrease after TAM treatment (MCF7 and T47D), whereas in ER& cells (SKBR3), no alterations in cell proliferation were observed, except for a small increase at 96 h. Karyotypes of both ER+ and ER& breast cancer cells increased in complexity after treatments with E2 and TAM leading to specific chromosomal abnormalities, some of which were consistent throughout the treatment duration. This genotoxic effect was higher in HER2+ cells. The ER&/HER2+ SKBR3 cells were found to be sensitive to TAM, exhibiting an increase in chromosomal aberrations. These in vitro results provide insights into the potential role of low doses of E2 and TAM in inducing chromosomal rearrangements in breast cancer cells.
Keywords: breast cancer cells; chromosomal abnormalities; chromosomal instability; estradiol; tamoxifen.
© 2016 Society for Endocrinology.
Figures





References
-
- Achuthan R, Bell SM, Roberts P, Leek JP, Horgan K, Markham AF, MacLennan KA, Speirs V. 2001. Genetic events during the transformation of a tamoxifen-sensitive human breast cancer cell line into a drug-resistant clone. Cancer Genetics and Cytogenetics 130 166–172. (10.1016/S0165-4608(01)00475-7) - DOI - PubMed
-
- Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, et al. 2001. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Research 61 8820–8829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

