Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome
- PMID: 27358031
- PMCID: PMC4928081
- DOI: 10.1038/srep28774
Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome
Abstract
Plant microbiome and its manipulation herald a new era for plant biotechnology with the potential to benefit sustainable crop production. However, studies evaluating the diversity, structure and impact of the microbiota in economic important crops are still rare. Here we describe a comprehensive inventory of the structure and assemblage of the bacterial and fungal communities associated with sugarcane. Our analysis identified 23,811 bacterial OTUs and an unexpected 11,727 fungal OTUs inhabiting the endophytic and exophytic compartments of roots, shoots, and leaves. These communities originate primarily from native soil around plants and colonize plant organs in distinct patterns. The sample type is the primary driver of fungal community assemblage, and the organ compartment plays a major role in bacterial community assemblage. We identified core bacterial and fungal communities composed of less than 20% of the total microbial richness but accounting for over 90% of the total microbial relative abundance. The roots showed 89 core bacterial families, 19 of which accounted for 44% of the total relative abundance. Stalks are dominated by groups of yeasts that represent over 12% of total relative abundance. The core microbiome described here comprise groups whose biological role underlies important traits in plant growth and fermentative processes.
Conflict of interest statement
Repsol, an oil company interested in bioenergy, financed this work.
Figures


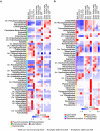
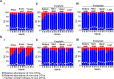


References
-
- Mendes R. et al.. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 (2011). - PubMed
-
- Lugtenberg B. & Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 (2009). - PubMed
-
- Hardoim P. R., van Overbeek L. S. & Elsas J. D. Van. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471 (2008). - PubMed
-
- Lebeis S. L. et al.. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

