The Variable Regions of Lactobacillus rhamnosus Genomes Reveal the Dynamic Evolution of Metabolic and Host-Adaptation Repertoires
- PMID: 27358423
- PMCID: PMC4943194
- DOI: 10.1093/gbe/evw123
The Variable Regions of Lactobacillus rhamnosus Genomes Reveal the Dynamic Evolution of Metabolic and Host-Adaptation Repertoires
Abstract
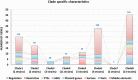
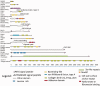
Lactobacillus rhamnosus is a diverse Gram-positive species with strains isolated from different ecological niches. Here, we report the genome sequence analysis of 40 diverse strains of L. rhamnosus and their genomic comparison, with a focus on the variable genome. Genomic comparison of 40 L. rhamnosus strains discriminated the conserved genes (core genome) and regions of plasticity involving frequent rearrangements and horizontal transfer (variome). The L. rhamnosus core genome encompasses 2,164 genes, out of 4,711 genes in total (the pan-genome). The accessory genome is dominated by genes encoding carbohydrate transport and metabolism, extracellular polysaccharides (EPS) biosynthesis, bacteriocin production, pili production, the cas system, and the associated clustered regularly interspaced short palindromic repeat (CRISPR) loci, and more than 100 transporter functions and mobile genetic elements like phages, plasmid genes, and transposons. A clade distribution based on amino acid differences between core (shared) proteins matched with the clade distribution obtained from the presence-absence of variable genes. The phylogenetic and variome tree overlap indicated that frequent events of gene acquisition and loss dominated the evolutionary segregation of the strains within this species, which is paralleled by evolutionary diversification of core gene functions. The CRISPR-Cas system could have contributed to this evolutionary segregation. Lactobacillus rhamnosus strains contain the genetic and metabolic machinery with strain-specific gene functions required to adapt to a large range of environments. A remarkable congruency of the evolutionary relatedness of the strains' core and variome functions, possibly favoring interspecies genetic exchanges, underlines the importance of gene-acquisition and loss within the L. rhamnosus strain diversification.
Keywords: comparative genomics; core and pan-genome; diversity; niche adaptation; probiotic.
© The Author(s) 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity.PLoS One. 2013 Jul 19;8(7):e68731. doi: 10.1371/journal.pone.0068731. Print 2013. PLoS One. 2013. PMID: 23894338 Free PMC article.
-
A large-scale comparative genomics study reveals niche-driven and within-sample intra-species functional diversification in Lacticaseibacillus rhamnosus.Food Res Int. 2023 Nov;173(Pt 2):113446. doi: 10.1016/j.foodres.2023.113446. Epub 2023 Sep 10. Food Res Int. 2023. PMID: 37803772
-
Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG.PLoS Genet. 2013;9(8):e1003683. doi: 10.1371/journal.pgen.1003683. Epub 2013 Aug 15. PLoS Genet. 2013. PMID: 23966868 Free PMC article.
-
[Why so rare if so essentiel: the determinants of the sparse distribution of CRISPR-Cas systems in bacterial genomes].Biol Aujourdhui. 2017;211(4):255-264. doi: 10.1051/jbio/2018005. Epub 2018 Jun 29. Biol Aujourdhui. 2017. PMID: 29956652 Review. French.
-
Role of CRISPR/cas system in the development of bacteriophage resistance.Adv Virus Res. 2012;82:289-338. doi: 10.1016/B978-0-12-394621-8.00011-X. Adv Virus Res. 2012. PMID: 22420856 Review.
Cited by
-
Comparative analysis of Lactobacillus gasseri from Chinese subjects reveals a new species-level taxa.BMC Genomics. 2020 Feb 3;21(1):119. doi: 10.1186/s12864-020-6527-y. BMC Genomics. 2020. PMID: 32013858 Free PMC article.
-
Ribosomal protein L4 of Lactobacillus rhamnosus LRB alters resistance to macrolides and other antibiotics.Mol Oral Microbiol. 2020 Jun;35(3):106-119. doi: 10.1111/omi.12281. Epub 2020 Feb 21. Mol Oral Microbiol. 2020. PMID: 32022979 Free PMC article.
-
Exploring the role of gut microbiota in antibiotic resistance and prevention.Ann Med. 2025 Dec;57(1):2478317. doi: 10.1080/07853890.2025.2478317. Epub 2025 Mar 17. Ann Med. 2025. PMID: 40096354 Free PMC article. Review.
-
[Research Progress in the Relationship Between Lactobacillus and Dental Caries].Sichuan Da Xue Xue Bao Yi Xue Ban. 2022 Sep;53(5):929-934. doi: 10.12182/20220960103. Sichuan Da Xue Xue Bao Yi Xue Ban. 2022. PMID: 36224699 Free PMC article. Review. Chinese.
-
The production and biochemical characterization of α-carbonic anhydrase from Lactobacillus rhamnosus GG.Appl Microbiol Biotechnol. 2022 Jun;106(11):4065-4074. doi: 10.1007/s00253-022-11990-3. Epub 2022 May 25. Appl Microbiol Biotechnol. 2022. PMID: 35612631 Free PMC article.
References
-
- Abedon ST. 2009. Phage evolution and ecology. Adv Appl Microbiol. 67:1–45. - PubMed
-
- Alemayehu D, et al. 2009. Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene 448:29–39. - PubMed
-
- Altermann E, et al. 1999. Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage ϕadh. Gene 236:333–346. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

