Single-cell analysis of glandular T cell receptors in Sjögren's syndrome
- PMID: 27358913
- PMCID: PMC4922426
- DOI: 10.1172/jci.insight.85609
Single-cell analysis of glandular T cell receptors in Sjögren's syndrome
Abstract
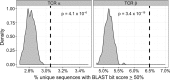
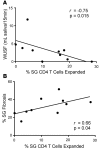
CD4+ T cells predominate in salivary gland (SG) inflammatory lesions in Sjögren's syndrome (SS). However, their antigen specificity, degree of clonal expansion, and relationship to clinical disease features remain unknown. We used multiplex reverse-transcriptase PCR to amplify paired T cell receptor α (TCRα) and β transcripts of single CD4+CD45RA- T cells from SG and peripheral blood (PB) of 10 individuals with primary SS, 9 of whom shared the HLA DR3/DQ2 risk haplotype. TCRα and β sequences were obtained from a median of 91 SG and 107 PB cells per subject. The degree of clonal expansion and frequency of cells expressing two productively rearranged α genes were increased in SG versus PB. Expanded clones from SG exhibited complementary-determining region 3 (CDR3) sequence similarity both within and among subjects, suggesting antigenic selection and shared antigen recognition. CDR3 similarities were shared among expanded clones from individuals discordant for canonical Ro and La autoantibodies, suggesting recognition of alternative SG antigen(s). The extent of SG clonal expansion correlated with reduced saliva production and increased SG fibrosis, linking expanded SG T cells with glandular dysfunction. Knowledge of paired TCRα and β sequences enables further work toward identification of target antigens and development of novel therapies.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

