The nature of mutations induced by replication–transcription collisions
- PMID: 27362223
- PMCID: PMC4945378
- DOI: 10.1038/nature18316
The nature of mutations induced by replication–transcription collisions
Abstract
The DNA replication and transcription machineries share a common DNA template and thus can collide with each other co-directionally or head-on. Replication–transcription collisions can cause replication fork arrest, premature transcription termination, DNA breaks, and recombination intermediates threatening genome integrity. Collisions may also trigger mutations, which are major contributors to genetic disease and evolution. However, the nature and mechanisms of collision-induced mutagenesis remain poorly understood. Here we reveal the genetic consequences of replication–transcription collisions in actively dividing bacteria to be two classes of mutations: duplications/deletions and base substitutions in promoters. Both signatures are highly deleterious but are distinct from the previously well-characterized base substitutions in the coding sequence. Duplications/deletions are probably caused by replication stalling events that are triggered by collisions; their distribution patterns are consistent with where the fork first encounters a transcription complex upon entering a transcription unit. Promoter substitutions result mostly from head-on collisions and frequently occur at a nucleotide that is conserved in promoters recognized by the major σ factor in bacteria. This substitution is generated via adenine deamination on the template strand in the promoter open complex, as a consequence of head-on replication perturbing transcription initiation. We conclude that replication–transcription collisions induce distinct mutation signatures by antagonizing replication and transcription, not only in coding sequences but also in gene regulatory elements.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






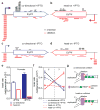

References
-
- French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–5. - PubMed
-
- Liu B, Alberts BM. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science. 1995;267:1131–7. - PubMed
-
- Vilette D, Ehrlich SD, Michel B. Transcription-induced deletions in Escherichia coli plasmids. Mol Microbiol. 1995;17:493–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

