Ki-67 acts as a biological surfactant to disperse mitotic chromosomes
- PMID: 27362226
- PMCID: PMC4947524
- DOI: 10.1038/nature18610
Ki-67 acts as a biological surfactant to disperse mitotic chromosomes
Abstract
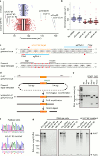
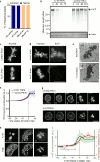
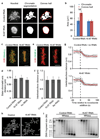
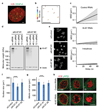
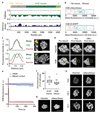
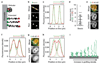
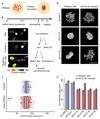
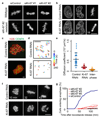
Eukaryotic genomes are partitioned into chromosomes that form compact and spatially well-separated mechanical bodies during mitosis. This enables chromosomes to move independently of each other for segregation of precisely one copy of the genome to each of the nascent daughter cells. Despite insights into the spatial organization of mitotic chromosomes and the discovery of proteins at the chromosome surface, the molecular and biophysical bases of mitotic chromosome structural individuality have remained unclear. Here we report that the proliferation marker protein Ki-67 (encoded by the MKI67 gene), a component of the mitotic chromosome periphery, prevents chromosomes from collapsing into a single chromatin mass after nuclear envelope disassembly, thus enabling independent chromosome motility and efficient interactions with the mitotic spindle. The chromosome separation function of human Ki-67 is not confined within a specific protein domain, but correlates with size and net charge of truncation mutants that apparently lack secondary structure. This suggests that Ki-67 forms a steric and electrostatic charge barrier, similar to surface-active agents (surfactants) that disperse particles or phase-separated liquid droplets in solvents. Fluorescence correlation spectroscopy showed a high surface density of Ki-67 and dual-colour labelling of both protein termini revealed an extended molecular conformation, indicating brush-like arrangements that are characteristic of polymeric surfactants. Our study thus elucidates a biomechanical role of the mitotic chromosome periphery in mammalian cells and suggests that natural proteins can function as surfactants in intracellular compartmentalization.
Figures



Comment in
-
Cell division: A sticky problem for chromosomes.Nature. 2016 Jul 14;535(7611):234-5. doi: 10.1038/nature18904. Epub 2016 Jun 29. Nature. 2016. PMID: 27362225 No abstract available.
-
Chromosome biology: Keeping chromosomes apart.Nat Rev Mol Cell Biol. 2016 Jul 21;17(8):462. doi: 10.1038/nrm.2016.95. Nat Rev Mol Cell Biol. 2016. PMID: 27440311 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

