Regulation of the Na+/K+-ATPase Ena1 Expression by Calcineurin/Crz1 under High pH Stress: A Quantitative Study
- PMID: 27362362
- PMCID: PMC4928930
- DOI: 10.1371/journal.pone.0158424
Regulation of the Na+/K+-ATPase Ena1 Expression by Calcineurin/Crz1 under High pH Stress: A Quantitative Study
Abstract
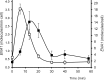
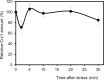
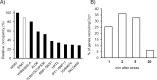
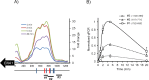
Regulated expression of the Ena1 Na+-ATPase is a crucial event for adaptation to high salt and/or alkaline pH stress in the budding yeast Saccharomyces cerevisiae. ENA1 expression is under the control of diverse signaling pathways, including that mediated by the calcium-regulatable protein phosphatase calcineurin and its downstream transcription factor Crz1. We present here a quantitative study of the expression of Ena1 in response to alkalinization of the environment and we analyze the contribution of Crz1 to this response. Experimental data and mathematical models substantiate the existence of two stress-responsive Crz1-binding sites in the ENA1 promoter and estimate that the contribution of Crz1 to the early response of the ENA1 promoter is about 60%. The models suggest the existence of a second input with similar kinetics, which would be likely mediated by high pH-induced activation of the Snf1 kinase.
Conflict of interest statement
Figures


References
-
- Banuelos MA, Sychrova H, Bleykasten-Grosshans C, Souciet JL, Potier S. The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 1998;144 (Pt 10)2749–58 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

