Crizotinib inhibits NF2-associated schwannoma through inhibition of focal adhesion kinase 1
- PMID: 27363027
- PMCID: PMC5342359
- DOI: 10.18632/oncotarget.10248
Crizotinib inhibits NF2-associated schwannoma through inhibition of focal adhesion kinase 1
Abstract
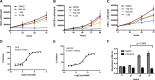
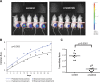
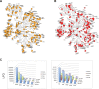
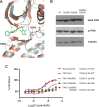
Neurofibromatosis type 2 (NF2) is a dominantly inherited autosomal disease characterized by schwannomas of the 8th cranial nerve. The NF2 tumor suppressor gene encodes for Merlin, a protein implicated as a suppressor of multiple cellular signaling pathways. To identify potential drug targets in NF2-associated malignancies we assessed the consequences of inhibiting the tyrosine kinase receptor MET. We identified crizotinib, a MET and ALK inhibitor, as a potent inhibitor of NF2-null Schwann cell proliferation in vitro and tumor growth in vivo. To identify the target/s of crizotnib we employed activity-based protein profiling (ABPP), leading to identification of FAK1 (PTK2) as the relevant target of crizotinib inhibition in NF2-null schwannoma cells. Subsequent studies confirm that inhibition of FAK1 is sufficient to suppress tumorigenesis in animal models of NF2 and that crizotinib-resistant forms of FAK1 can rescue the effects of treatment. These studies identify a FDA approved drug as a potential treatment for NF2 and delineate the mechanism of action in NF2-null Schwann cells.
Keywords: FAK; NF2; crizotinib; neurofibromatosis; signal transduction.
Conflict of interest statement
The authors declare no conflicts of interests.
Figures

References
-
- Gusella JF, Ramesh V, MacCollin M, Jacoby LB. Merlin: the neurofibromatosis 2 tumor suppressor. Biochim Biophys Acta. 1999;1423:M29–36. - PubMed
-
- Torres-Martin M, Lassaletta L, San-Roman-Montero J, De Campos JM, Isla A, Gavilan J, Melendez B, Pinto GR, Burbano RR, Castresana JS, Rey JA. Microarray analysis of gene expression in vestibular schwannomas reveals SPP1/MET signaling pathway and androgen receptor deregulation. Int J Oncol. 2013 - PMC - PubMed
-
- Kissil JL, Wilker EW, Johnson KC, Eckman MS, Yaffe MB, Jacks T. Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol Cell. 2003;12:841–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

