PhytoCRISP-Ex: a web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing
- PMID: 27363443
- PMCID: PMC4929763
- DOI: 10.1186/s12859-016-1143-1
PhytoCRISP-Ex: a web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing
Abstract
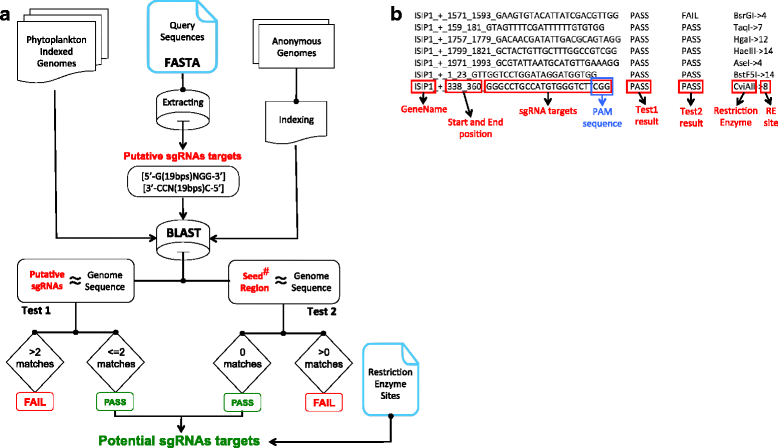
Background: With the emerging interest in phytoplankton research, the need to establish genetic tools for the functional characterization of genes is indispensable. The CRISPR/Cas9 system is now well recognized as an efficient and accurate reverse genetic tool for genome editing. Several computational tools have been published allowing researchers to find candidate target sequences for the engineering of the CRISPR vectors, while searching possible off-targets for the predicted candidates. These tools provide built-in genome databases of common model organisms that are used for CRISPR target prediction. Although their predictions are highly sensitive, the applicability to non-model genomes, most notably protists, makes their design inadequate. This motivated us to design a new CRISPR target finding tool, PhytoCRISP-Ex. Our software offers CRIPSR target predictions using an extended list of phytoplankton genomes and also delivers a user-friendly standalone application that can be used for any genome.
Results: The software attempts to integrate, for the first time, most available phytoplankton genomes information and provide a web-based platform for Cas9 target prediction within them with high sensitivity. By offering a standalone version, PhytoCRISP-Ex maintains an independence to be used with any organism and widens its applicability in high throughput pipelines. PhytoCRISP-Ex out pars all the existing tools by computing the availability of restriction sites over the most probable Cas9 cleavage sites, which can be ideal for mutant screens.
Conclusions: PhytoCRISP-Ex is a simple, fast and accurate web interface with 13 pre-indexed and presently updating phytoplankton genomes. The software was also designed as a UNIX-based standalone application that allows the user to search for target sequences in the genomes of a variety of other species.
Keywords: CRISPR; Cas9; Eukaryotes; Genome editing; Protists.
Figures
References
-
- Aach JEA. CasFinder: Flexible algorithm for identifying specific Cas9 targets in genomes. bioRxiv. (2014);005074. doi:http://dx.doi.org/10.1101/005074. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

