Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data
- PMID: 27363486
- PMCID: PMC4929559
- DOI: 10.1038/srep29033
Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data
Abstract
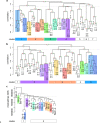
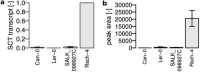
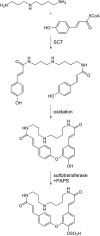
Many metabolomics studies focus on aboveground parts of the plant, while metabolism within roots and the chemical composition of the rhizosphere, as influenced by exudation, are not deeply investigated. In this study, we analysed exudate metabolic patterns of Arabidopsis thaliana and their variation in genetically diverse accessions. For this project, we used the 19 parental accessions of the Arabidopsis MAGIC collection. Plants were grown in a hydroponic system, their exudates were harvested before bolting and subjected to UPLC/ESI-QTOF-MS analysis. Metabolite profiles were analysed together with the genome sequence information. Our study uncovered distinct metabolite profiles for root exudates of the 19 accessions. Hierarchical clustering revealed similarities in the exudate metabolite profiles, which were partly reflected by the genetic distances. An association of metabolite absence with nonsense mutations was detected for the biosynthetic pathways of an indolic glucosinolate hydrolysis product, a hydroxycinnamic acid amine and a flavonoid triglycoside. Consequently, a direct link between metabolic phenotype and genotype was detected without using segregating populations. Moreover, genomics can help to identify biosynthetic enzymes in metabolomics experiments. Our study elucidates the chemical composition of the rhizosphere and its natural variation in A. thaliana, which is important for the attraction and shaping of microbial communities.
Figures



References
-
- Mithen R., Clarke J. H., Lister C. & Dean C. Genetics of aliphatic glucosinolates. III. Side chain structure of aliphatic glucosinolates in Arabidopsis thaliana. Heredity (Edinb). 74, 210–215 (1995).
-
- Mithen R. & Campos H. Genetic variation of aliphatic glucosinolates in Arabidopsis thaliana and prospects for map based gene cloning. Entomologica Experimentalis et Applicanta. 53, 202–205 (1996).
-
- Magrath R. et al. Genetics of aliphatic glucosinolates. I. Side chain elongation in Brassica napus and Arabidopsis thaliana. Heredity (Edinb). 72, 290–299 (1994).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

