CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing
- PMID: 27363488
- PMCID: PMC4929496
- DOI: 10.1038/srep28973
CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing
Abstract
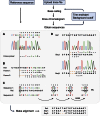
The advent of next generation gene editing technologies has revolutionized the fields of genome engineering in allowing the generation of gene knockout models and functional gene analysis. However, the screening of resultant clones remains challenging due to the simultaneous presence of different indels. Here, we present CRISP-ID, a web application which uses a unique algorithm for genotyping up to three alleles from a single Sanger sequencing trace, providing a robust and readily accessible platform to directly identify indels and significantly speed up the characterization of clones.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Urnov F. D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

