Specificity and mechanism of action of alpha-helical membrane-active peptides interacting with model and biological membranes by single-molecule force spectroscopy
- PMID: 27363513
- PMCID: PMC4929710
- DOI: 10.1038/srep29145
Specificity and mechanism of action of alpha-helical membrane-active peptides interacting with model and biological membranes by single-molecule force spectroscopy
Abstract
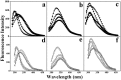
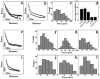
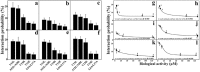
In this study, to systematically investigate the targeting specificity of membrane-active peptides on different types of cell membranes, we evaluated the effects of peptides on different large unilamellar vesicles mimicking prokaryotic, normal eukaryotic, and cancer cell membranes by single-molecule force spectroscopy and spectrum technology. We revealed that cationic membrane-active peptides can exclusively target negatively charged prokaryotic and cancer cell model membranes rather than normal eukaryotic cell model membranes. Using Acholeplasma laidlawii, 3T3-L1, and HeLa cells to represent prokaryotic cells, normal eukaryotic cells, and cancer cells in atomic force microscopy experiments, respectively, we further studied that the single-molecule targeting interaction between peptides and biological membranes. Antimicrobial and anticancer activities of peptides exhibited strong correlations with the interaction probability determined by single-molecule force spectroscopy, which illustrates strong correlations of peptide biological activities and peptide hydrophobicity and charge. Peptide specificity significantly depends on the lipid compositions of different cell membranes, which validates the de novo design of peptide therapeutics against bacteria and cancers.
Figures
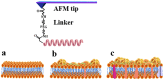
Similar articles
-
The study of single anticancer peptides interacting with HeLa cell membranes by single molecule force spectroscopy.Nanoscale. 2012 Feb 21;4(4):1283-6. doi: 10.1039/c2nr11541g. Epub 2012 Jan 3. Nanoscale. 2012. PMID: 22215262
-
Cationic peptide-induced remodelling of model membranes: direct visualization by in situ atomic force microscopy.J Struct Biol. 2008 Apr;162(1):121-38. doi: 10.1016/j.jsb.2007.11.003. Epub 2007 Nov 17. J Struct Biol. 2008. PMID: 18180166
-
Peptide hydrophobicity controls the activity and selectivity of magainin 2 amide in interaction with membranes.Biochemistry. 1997 May 20;36(20):6124-32. doi: 10.1021/bi9619987. Biochemistry. 1997. PMID: 9166783
-
Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells.Biochim Biophys Acta. 1999 Dec 15;1462(1-2):71-87. doi: 10.1016/s0005-2736(99)00201-1. Biochim Biophys Acta. 1999. PMID: 10590303 Review.
-
How Membrane-Active Peptides Get into Lipid Membranes.Acc Chem Res. 2016 Jun 21;49(6):1130-8. doi: 10.1021/acs.accounts.6b00074. Epub 2016 May 17. Acc Chem Res. 2016. PMID: 27187572 Review.
Cited by
-
Fluorescence Imaging of Disrupted Interfaces between Liquid-Ordered and Liquid-Disordered Domains by a Flavin-Labeled PNA Duplex.ACS Omega. 2017 Jun 22;2(6):2912-2915. doi: 10.1021/acsomega.7b00581. eCollection 2017 Jun 30. ACS Omega. 2017. PMID: 31457626 Free PMC article.
-
Enantiomeric Effect of d-Amino Acid Substitution on the Mechanism of Action of α-Helical Membrane-Active Peptides.Int J Mol Sci. 2017 Dec 27;19(1):67. doi: 10.3390/ijms19010067. Int J Mol Sci. 2017. PMID: 29280948 Free PMC article.
-
Peptide hemolytic activity analysis using visual data mining of similarity-based complex networks.NPJ Syst Biol Appl. 2024 Oct 4;10(1):115. doi: 10.1038/s41540-024-00429-2. NPJ Syst Biol Appl. 2024. PMID: 39367008 Free PMC article.
-
Exclusion of Anchor-Matched Peptide Nucleic Acid from Liquid-Ordered Domains by Hybridization with Complementary Flavin-Labeled DNA.ACS Omega. 2022 Dec 16;8(1):1109-1113. doi: 10.1021/acsomega.2c06463. eCollection 2023 Jan 10. ACS Omega. 2022. PMID: 36643542 Free PMC article.
-
Unravelling the molecular effect of ocellatin-1, F1, K1 and S1, the frog-skin antimicrobial peptides to enhance its therapeutics-quantum and molecular mechanical approaches.J Mol Model. 2021 Jan 3;27(1):10. doi: 10.1007/s00894-020-04652-6. J Mol Model. 2021. PMID: 33392722
References
-
- Dong N. et al. Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials 35, 8028–8039 (2014). - PubMed
-
- Hong J., Oren Z. & Shai Y. Structure and organization of hemolytic and nonhemolytic diastereomers of antimicrobial peptides in membranes. Biochemistry 38, 16963–16973 (1999). - PubMed
-
- Ehrenstein G. & Lecar H. Electrically gated ionic channels in lipid bilayers. Q. Rev. Biophys. 10, 1–34 (1977). - PubMed
-
- Shan Y. & Wang H. The structure and function of cell membranes examined by atomic force microscopy and single-molecule force spectroscopy. Chem. Soc. Rev. 44, 3617–3638 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

