Still and rotating myosin clusters determine cytokinetic ring constriction
- PMID: 27363521
- PMCID: PMC4932180
- DOI: 10.1038/ncomms11860
Still and rotating myosin clusters determine cytokinetic ring constriction
Abstract
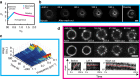
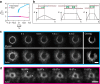
The cytokinetic ring is essential for separating daughter cells during division. It consists of actin filaments and myosin motors that are generally assumed to organize as sarcomeres similar to skeletal muscles. However, direct evidence is lacking. Here we show that the internal organization and dynamics of rings are different from sarcomeres and distinct in different cell types. Using micro-cavities to orient rings in single focal planes, we find in mammalian cells a transition from a homogeneous distribution to a periodic pattern of myosin clusters at the onset of constriction. In contrast, in fission yeast, myosin clusters rotate prior to and during constriction. Theoretical analysis indicates that both patterns result from acto-myosin self-organization and reveals differences in the respective stresses. These findings suggest distinct functional roles for rings: contraction in mammalian cells and transport in fission yeast. Thus self-organization under different conditions may be a generic feature for regulating morphogenesis in vivo.
Figures




References
-
- Rappaport R. Cytokinesis in Animal Cells Cambridge Univ. Press (1996).
-
- Schroeder T. E. Molecules and Cell Movement 305–334Raven Press (1975).
-
- Eggert U. S., Mitchison T. J. & Field C. M. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75, 543–566 (2006). - PubMed
-
- Carvalho A., Desai A. & Oegema K. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell 137, 926–937 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

