A next generation sequencing-based method to study the intra-host genetic diversity of norovirus in patients with acute and chronic infection
- PMID: 27363999
- PMCID: PMC4929757
- DOI: 10.1186/s12864-016-2831-y
A next generation sequencing-based method to study the intra-host genetic diversity of norovirus in patients with acute and chronic infection
Abstract
Background: Immunocompromised individuals with chronic norovirus (NoV) infection and elderly patients are hypothesized to be reservoirs where NoV might accumulate mutations and evolve into pandemic strains. Next generation sequencing (NGS) methods can monitor the intra-host diversity of NoV and its evolution but low abundance of viral RNA results in sub-optimal efficiency. In this study, we: 1) established a next generation sequencing-based method for NoV using bacterial rRNA depletion as a viral RNA enrichment strategy, and 2) measured the intra-host genetic diversity of NoV in specimens of patients with acute NoV infection (n = 4) and in longitudinal specimens of an immunocompromised patient with chronic NoV infection (n = 2).
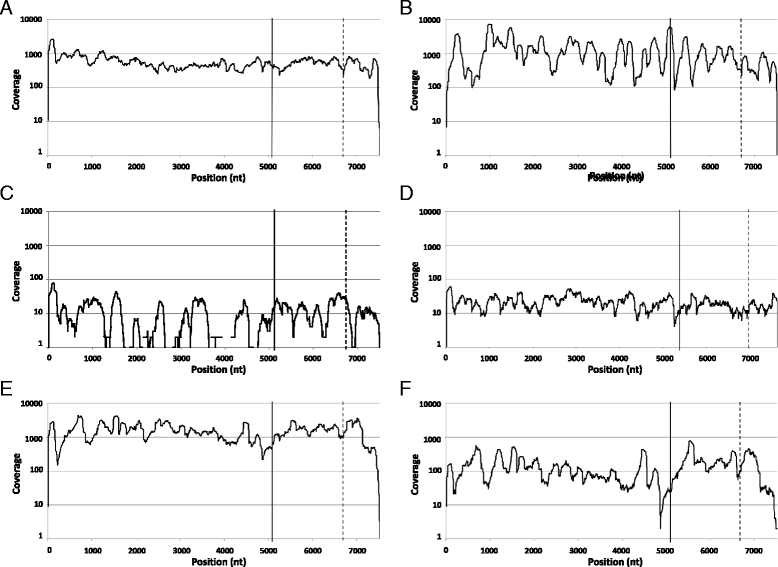
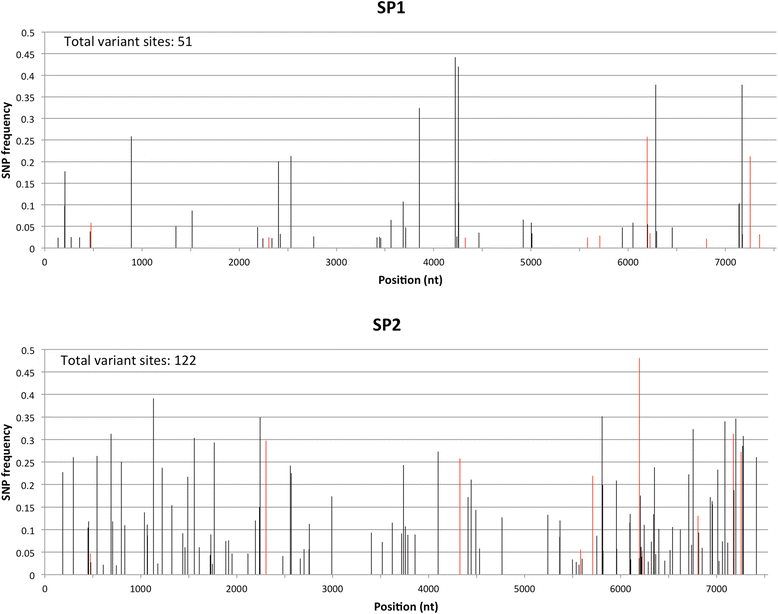
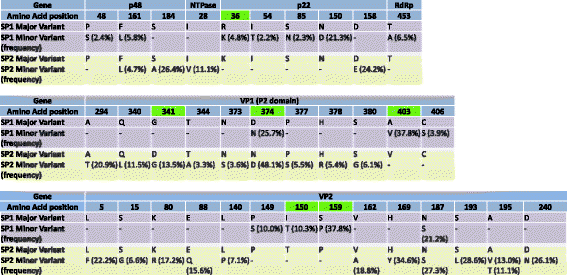
Results: A single Illumina MiSeq dataset resulted in near full-length genome sequences for 5 out of 6 multiplexed samples. Experimental depletion of bacterial rRNA in stool RNA provided up to 1.9 % of NoV reads. The intra-host viral population in patients with acute NoV infection was homogenous and no single nucleotide variants (SNVs) were detected. In contrast, the NoV population from the immunocompromised patient was highly diverse and accumulated SNVs over time (51 SNVs in the first sample and 122 SNVs in the second sample collected 4 months later). The percentages of SNVs causing non-synonymous mutations were 27.5 % and 20.5 % for the first and second samples, respectively. The majority of non-synonymous mutations occurred, in increasing order of frequency, in p22, the major capsid (VP1) and minor capsid (VP2) genes.
Conclusions: The results provide data useful for the selection and improvement of NoV RNA enrichment strategies for NGS. Whole genome analysis using next generation sequencing confirmed that the within-host population of NoV in an immunocompromised individual with chronic NoV infection was more diverse compared to that in individuals with acute infection. We also observed an accumulation of non-synonymous mutations at the minor capsid gene that has not been reported in previous studies and might have a role in NoV adaptation.
Keywords: Gastroenteritis outbreaks; Genetic variation; Immunocompromised host; Molecular evolution; Next generation sequencing; Norovirus.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

