Activation of Wnt/β-Catenin in Ewing Sarcoma Cells Antagonizes EWS/ETS Function and Promotes Phenotypic Transition to More Metastatic Cell States
- PMID: 27364557
- PMCID: PMC5010452
- DOI: 10.1158/0008-5472.CAN-15-3422
Activation of Wnt/β-Catenin in Ewing Sarcoma Cells Antagonizes EWS/ETS Function and Promotes Phenotypic Transition to More Metastatic Cell States
Abstract
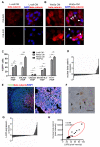
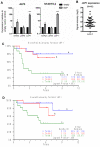
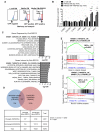
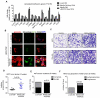
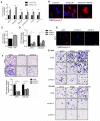
Ewing sarcomas are characterized by the presence of EWS/ETS fusion genes in the absence of other recurrent genetic alterations and mechanisms of tumor heterogeneity that contribute to disease progression remain unclear. Mutations in the Wnt/β-catenin pathway are rare in Ewing sarcoma but the Wnt pathway modulator LGR5 is often highly expressed, suggesting a potential role for the axis in tumor pathogenesis. We evaluated β-catenin and LGR5 expression in Ewing sarcoma cell lines and tumors and noted marked intra- and inter-tumor heterogeneity. Tumors with evidence of active Wnt/β-catenin signaling were associated with increased incidence of tumor relapse and worse overall survival. Paradoxically, RNA sequencing revealed a marked antagonism of EWS/ETS transcriptional activity in Wnt/β-catenin-activated tumor cells. Consistent with this, Wnt/β-catenin-activated cells displayed a phenotype that was reminiscent of Ewing sarcoma cells with partial EWS/ETS loss of function. Specifically, activation of Wnt/β-catenin induced alterations to the actin cytoskeleton, acquisition of a migratory phenotype, and upregulation of EWS/ETS-repressed genes. Notably, activation of Wnt/β-catenin signaling led to marked induction of tenascin C (TNC), an established promoter of cancer metastasis, and an EWS/ETS-repressed target gene. Loss of TNC function in Ewing sarcoma cells profoundly inhibited their migratory and metastatic potential. Our studies reveal that heterogeneous activation of Wnt/β-catenin signaling in subpopulations of tumor cells contributes to phenotypic heterogeneity and disease progression in Ewing sarcoma. Significantly, this is mediated, at least in part, by inhibition of EWS/ETS fusion protein function that results in derepression of metastasis-associated gene programs. Cancer Res; 76(17); 5040-53. ©2016 AACR.
©2016 American Association for Cancer Research.
Figures

References
-
- Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer discovery. 2014 Nov;4(11):1326–41. PubMed PMID: 25186949. Epub 2014/09/05. eng. - PubMed
-
- Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer discovery. 2014 Nov;4(11):1342–53. PubMed PMID: 25223734. Pubmed Central PMCID: 4264969. Epub 2014/09/17. eng. - PMC - PubMed
-
- Bennani-Baiti IM, Cooper A, Lawlor ER, Kauer M, Ban J, Aryee DN, et al. Intercohort gene expression co-analysis reveals chemokine receptors as prognostic indicators in Ewing’s sarcoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010 Jul 15;16(14):3769–78. PubMed PMID: 20525755. Pubmed Central PMCID: 2905506. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

