Oligonucleotide-based systems: DNA, microRNAs, DNA/RNA aptamers
- PMID: 27365033
- PMCID: PMC4986462
- DOI: 10.1042/EBC20150004
Oligonucleotide-based systems: DNA, microRNAs, DNA/RNA aptamers
Abstract
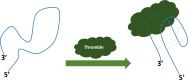
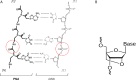
There are an increasing number of applications that have been developed for oligonucleotide-based biosensing systems in genetics and biomedicine. Oligonucleotide-based biosensors are those where the probe to capture the analyte is a strand of deoxyribonucleic acid (DNA), ribonucleic acid (RNA) or a synthetic analogue of naturally occurring nucleic acids. This review will shed light on various types of nucleic acids such as DNA and RNA (particularly microRNAs), their role and their application in biosensing. It will also cover DNA/RNA aptamers, which can be used as bioreceptors for a wide range of targets such as proteins, small molecules, bacteria and even cells. It will also highlight how the invention of synthetic oligonucleotides such as peptide nucleic acid (PNA) or locked nucleic acid (LNA) has pushed the limits of molecular biology and biosensor development to new perspectives. These technologies are very promising albeit still in need of development in order to bridge the gap between the laboratory-based status and the reality of biomedical applications.
Keywords: aptamers; biosensing; locked nucleic acids; microRNAs; oligonucleotides; peptide nucleic acids.
© 2016 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

