Determination of half-maximal inhibitory concentration using biosensor-based protein interaction analysis
- PMID: 27365221
- PMCID: PMC4955526
- DOI: 10.1016/j.ab.2016.06.025
Determination of half-maximal inhibitory concentration using biosensor-based protein interaction analysis
Abstract
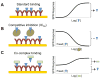
Half-maximal inhibitory concentration (IC50) is the most widely used and informative measure of a drug's efficacy. It indicates how much drug is needed to inhibit a biological process by half, thus providing a measure of potency of an antagonist drug in pharmacological research. Most approaches to determine IC50 of a pharmacological compound are based on assays that utilize whole cell systems. While they generally provide outstanding potency information, results can depend on the experimental cell line used and may not differentiate a compound's ability to inhibit specific interactions. Here we show using the secreted Transforming Growth Factor-β (TGF-β) family ligand BMP-4 and its receptors as example that surface plasmon resonance can be used to accurately determine IC50 values of individual ligand-receptor pairings. The molecular resolution achievable wih this approach can help distinguish inhibitors that specifically target individual complexes, or that can inhibit multiple functional interactions at the same time.
Keywords: BMP-4; Bone morphogenetic protein; Cerberus; IC(50); Inhibitor; SPR; Surface plasmon resonance; TGF-β.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- LeRoith D, Bondy C. Growth factors and cytokines in health and disease : a multi-volume treatise. JAI Press; Greenwich, Conn: 1996.
-
- Vilcek J, Feldmann M. Historical review: Cytokines as therapeutics and targets of therapeutics. Trends in pharmacological sciences. 2004;25:201–209. - PubMed
-
- Oppenheim JJ. Cytokines: past, present, and future. International journal of hematology. 2001;74:3–8. - PubMed
-
- Elvin JG, Couston RG, van der Walle CF. Therapeutic antibodies: market considerations, disease targets and bioprocessing. International journal of pharmaceutics. 2013;440:83–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

