Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene
- PMID: 27365461
- PMCID: PMC5004465
- DOI: 10.3324/haematol.2015.135475
Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene
Abstract
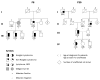
Hodgkin lymphoma shows strong familial aggregation but no major susceptibility genes have been identified to date. The goal of this study was to identify high-penetrance variants using whole exome sequencing in 17 Hodgkin lymphoma prone families with three or more affected cases or obligate carriers (69 individuals), followed by targeted sequencing in an additional 48 smaller HL families (80 individuals). Alignment and variant calling were performed using standard methods. Dominantly segregating, rare, coding or potentially functional variants were further prioritized based on predicted deleteriousness, conservation, and potential importance in lymphoid malignancy pathways. We selected 23 genes for targeted sequencing. Only the p.A1065T variant in KDR (kinase insert domain receptor) also known as VEGFR2 (vascular endothelial growth factor receptor 2) was replicated in two independent Hodgkin lymphoma families. KDR is a type III receptor tyrosine kinase, the main mediator of vascular endothelial growth factor induced proliferation, survival, and migration. Its activity is associated with several diseases including lymphoma. Functional experiments have shown that p.A1065T, located in the activation loop, can promote constitutive autophosphorylation on tyrosine in the absence of vascular endothelial growth factor and that the kinase activity was abrogated after exposure to kinase inhibitors. A few other promising mutations were identified but appear to be "private". In conclusion, in the largest sequenced cohort of Hodgkin lymphoma families to date, we identified a causal mutation in the KDR gene. While independent validation is needed, this mutation may increase downstream tumor cell proliferation activity and might be a candidate for targeted therapy.
Copyright© Ferrata Storti Foundation.
Figures

References
-
- Howlander N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2011, National Cancer Institute. 2014. [cited 2014 04/01/2014]; Available from: http://seer.cancer.gov/csr/1975_2011/
-
- Caporaso NE, Goldin LR, Anderson WF, Landgren O. Current insight on trends, causes, and mechanisms of Hodgkin’s lymphoma. Cancer J. 2009;15(2):117–123. - PubMed
-
- Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86(21):1600–1608. - PubMed
-
- Goldin LR, Pfeiffer RM, Gridley G, et al. Familial aggregation of Hodgkin lymphoma and related tumors. Cancer. 2004; 100(9):1902–1908. - PubMed
-
- Lindelof B, Eklund G. Analysis of hereditary component of cancer by use of a familial index by site. Lancet. 2001; 358(9294):1696–1698. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

