Cellular Interrogation: Exploiting Cell-to-Cell Variability to Discriminate Regulatory Mechanisms in Oscillatory Signalling
- PMID: 27367445
- PMCID: PMC4930170
- DOI: 10.1371/journal.pcbi.1004995
Cellular Interrogation: Exploiting Cell-to-Cell Variability to Discriminate Regulatory Mechanisms in Oscillatory Signalling
Abstract
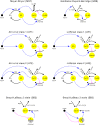
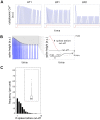
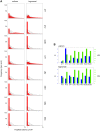
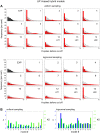
The molecular complexity within a cell may be seen as an evolutionary response to the external complexity of the cell's environment. This suggests that the external environment may be harnessed to interrogate the cell's internal molecular architecture. Cells, however, are not only nonlinear and non-stationary, but also exhibit heterogeneous responses within a clonal, isogenic population. In effect, each cell undertakes its own experiment. Here, we develop a method of cellular interrogation using programmable microfluidic devices which exploits the additional information present in cell-to-cell variation, without requiring model parameters to be fitted to data. We focussed on Ca2+ signalling in response to hormone stimulation, which exhibits oscillatory spiking in many cell types and chose eight models of Ca2+ signalling networks which exhibit similar behaviour in simulation. We developed a nonlinear frequency analysis for non-stationary responses, which could classify models into groups under parameter variation, but found that this question alone was unable to distinguish critical feedback loops. We further developed a nonlinear amplitude analysis and found that the combination of both questions ruled out six of the models as inconsistent with the experimentally-observed dynamics and heterogeneity. The two models that survived the double interrogation were mathematically different but schematically identical and yielded the same unexpected predictions that we confirmed experimentally. Further analysis showed that subtle mathematical details can markedly influence non-stationary responses under parameter variation, emphasising the difficulty of finding a "correct" model. By developing questions for the pathway being studied, and designing more versatile microfluidics, cellular interrogation holds promise as a systematic strategy that can complement direct intervention by genetics or pharmacology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

