Structure and Functional Diversity of GCN5-Related N-Acetyltransferases (GNAT)
- PMID: 27367672
- PMCID: PMC4964394
- DOI: 10.3390/ijms17071018
Structure and Functional Diversity of GCN5-Related N-Acetyltransferases (GNAT)
Abstract
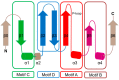
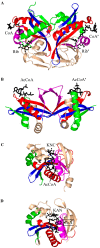
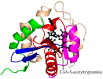
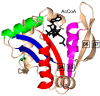
General control non-repressible 5 (GCN5)-related N-acetyltransferases (GNAT) catalyze the transfer of an acyl moiety from acyl coenzyme A (acyl-CoA) to a diverse group of substrates and are widely distributed in all domains of life. This review of the currently available data acquired on GNAT enzymes by a combination of structural, mutagenesis and kinetic methods summarizes the key similarities and differences between several distinctly different families within the GNAT superfamily, with an emphasis on the mechanistic insights obtained from the analysis of the complexes with substrates or inhibitors. It discusses the structural basis for the common acetyltransferase mechanism, outlines the factors important for the substrate recognition, and describes the mechanism of action of inhibitors of these enzymes. It is anticipated that understanding of the structural basis behind the reaction and substrate specificity of the enzymes from this superfamily can be exploited in the development of novel therapeutics to treat human diseases and combat emerging multidrug-resistant microbial infections.
Keywords: GNAT; acetyltransferase; catalytic residues; crystal structure; enzyme inhibitor; reaction mechanism.
Figures








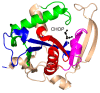
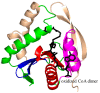

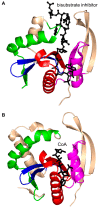
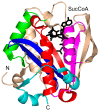

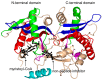

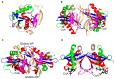


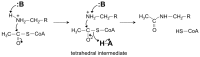
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

