Ribosomal Antibiotics: Contemporary Challenges
- PMID: 27367739
- PMCID: PMC5039520
- DOI: 10.3390/antibiotics5030024
Ribosomal Antibiotics: Contemporary Challenges
Abstract
Most ribosomal antibiotics obstruct distinct ribosomal functions. In selected cases, in addition to paralyzing vital ribosomal tasks, some ribosomal antibiotics are involved in cellular regulation. Owing to the global rapid increase in the appearance of multi-drug resistance in pathogenic bacterial strains, and to the extremely slow progress in developing new antibiotics worldwide, it seems that, in addition to the traditional attempts at improving current antibiotics and the intensive screening for additional natural compounds, this field should undergo substantial conceptual revision. Here, we highlight several contemporary issues, including challenging the common preference of broad-range antibiotics; the marginal attention to alterations in the microbiome population resulting from antibiotics usage, and the insufficient awareness of ecological and environmental aspects of antibiotics usage. We also highlight recent advances in the identification of species-specific structural motifs that may be exploited for the design and the creation of novel, environmental friendly, degradable, antibiotic types, with a better distinction between pathogens and useful bacterial species in the microbiome. Thus, these studies are leading towards the design of "pathogen-specific antibiotics," in contrast to the current preference of broad range antibiotics, partially because it requires significant efforts in speeding up the discovery of the unique species motifs as well as the clinical pathogen identification.
Keywords: microbiome; multi-drug resistance; novel degradable antibiotics; species-specific antibiotics susceptibility.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

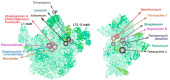

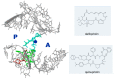


References
-
- Gibbons A. Resistance to antibiotics found in isolated Amazonian tribe. Science AAAS. 2015 doi: 10.1126/science.aab2509. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

