GiniClust: detecting rare cell types from single-cell gene expression data with Gini index
- PMID: 27368803
- PMCID: PMC4930624
- DOI: 10.1186/s13059-016-1010-4
GiniClust: detecting rare cell types from single-cell gene expression data with Gini index
Abstract
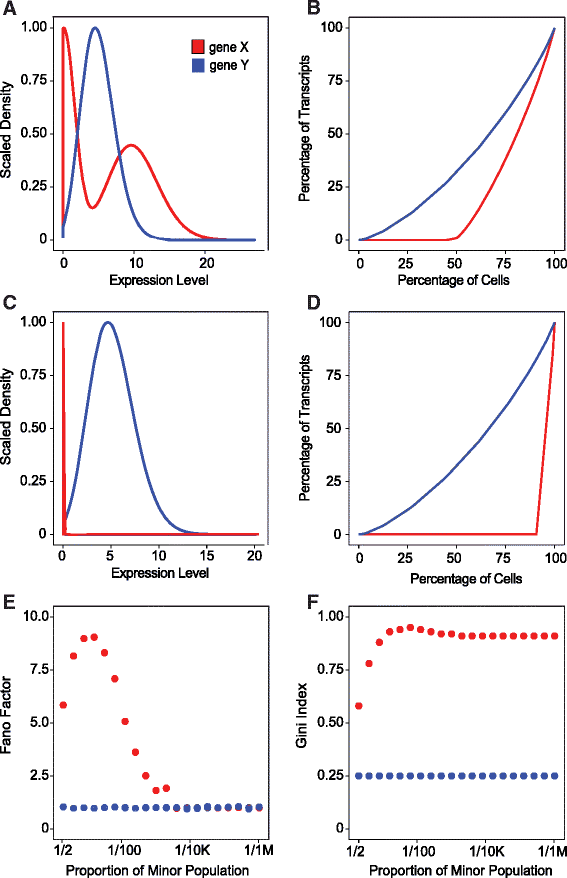
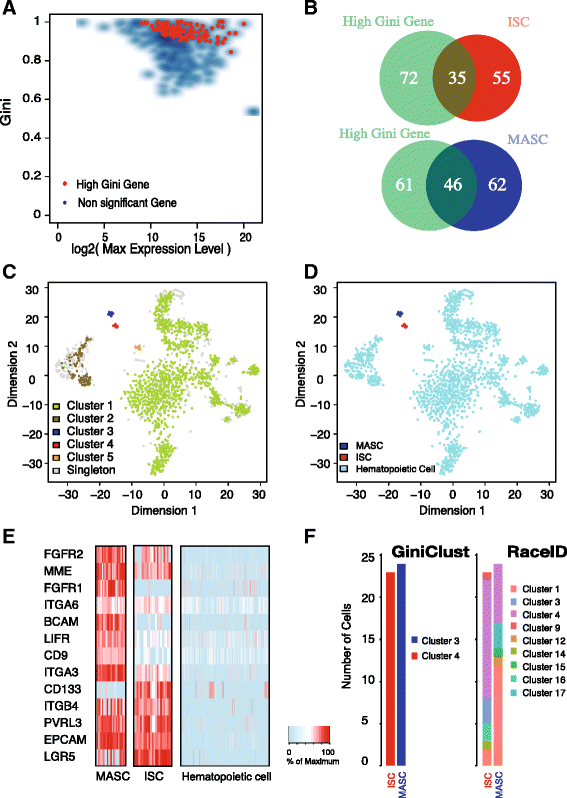
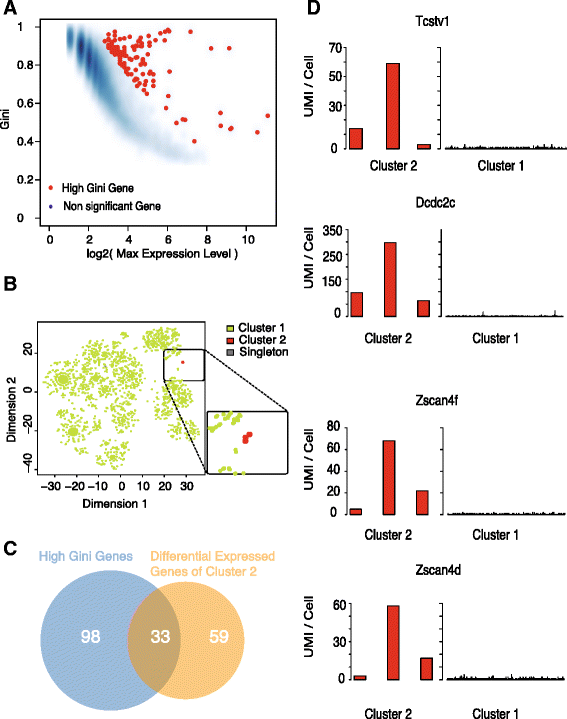
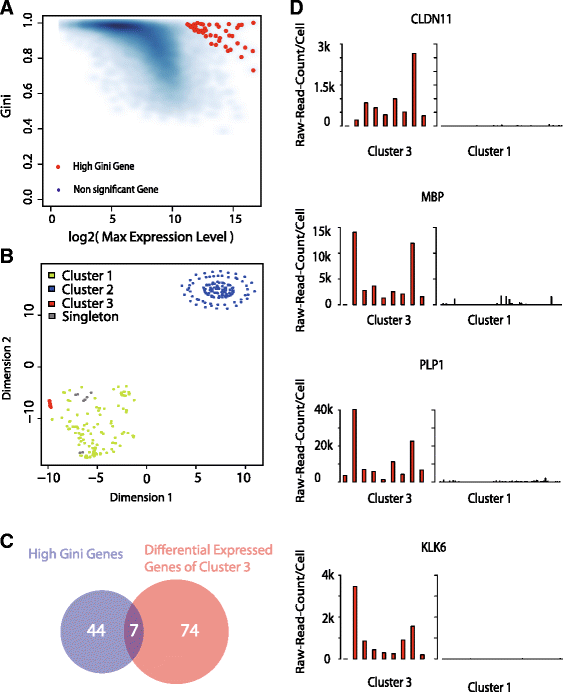
High-throughput single-cell technologies have great potential to discover new cell types; however, it remains challenging to detect rare cell types that are distinct from a large population. We present a novel computational method, called GiniClust, to overcome this challenge. Validation against a benchmark dataset indicates that GiniClust achieves high sensitivity and specificity. Application of GiniClust to public single-cell RNA-seq datasets uncovers previously unrecognized rare cell types, including Zscan4-expressing cells within mouse embryonic stem cells and hemoglobin-expressing cells in the mouse cortex and hippocampus. GiniClust also correctly detects a small number of normal cells that are mixed in a cancer cell population.
Keywords: Clustering; Gini index; RNA-seq; Rare cell type; Single-cell analysis; qPCR.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

