A basic domain in the histone H2B N-terminal tail is important for nucleosome assembly by FACT
- PMID: 27369377
- PMCID: PMC5100577
- DOI: 10.1093/nar/gkw588
A basic domain in the histone H2B N-terminal tail is important for nucleosome assembly by FACT
Abstract
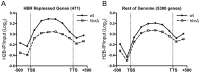
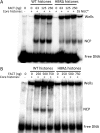
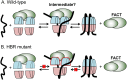
Nucleosome assembly in vivo requires assembly factors, such as histone chaperones, to bind to histones and mediate their deposition onto DNA. In yeast, the essential histone chaperone FACT (FAcilitates Chromatin Transcription) functions in nucleosome assembly and H2A-H2B deposition during transcription elongation and DNA replication. Recent studies have identified candidate histone residues that mediate FACT binding to histones, but it is not known which histone residues are important for FACT to deposit histones onto DNA during nucleosome assembly. In this study, we report that the histone H2B repression (HBR) domain within the H2B N-terminal tail is important for histone deposition by FACT. Deletion of the HBR domain causes significant defects in histone occupancy in the yeast genome, particularly at HBR-repressed genes, and a pronounced increase in H2A-H2B dimers that remain bound to FACT in vivo Moreover, the HBR domain is required for purified FACT to efficiently assemble recombinant nucleosomes in vitro We propose that the interaction between the highly basic HBR domain and DNA plays an important role in stabilizing the nascent nucleosome during the process of histone H2A-H2B deposition by FACT.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Residues in the Nucleosome Acidic Patch Regulate Histone Occupancy and Are Important for FACT Binding in Saccharomyces cerevisiae.Genetics. 2017 Jul;206(3):1339-1348. doi: 10.1534/genetics.117.201939. Epub 2017 May 3. Genetics. 2017. PMID: 28468903 Free PMC article.
-
[Structure and function of histone chaperone FACT].Mol Biol (Mosk). 2015 Nov-Dec;49(6):891-904. doi: 10.7868/S0026898415060026. Mol Biol (Mosk). 2015. PMID: 26710768 Review. Russian.
-
The Abundant Histone Chaperones Spt6 and FACT Collaborate to Assemble, Inspect, and Maintain Chromatin Structure in Saccharomyces cerevisiae.Genetics. 2015 Nov;201(3):1031-45. doi: 10.1534/genetics.115.180794. Epub 2015 Sep 28. Genetics. 2015. PMID: 26416482 Free PMC article.
-
Insight into the mechanism of nucleosome reorganization from histone mutants that suppress defects in the FACT histone chaperone.Genetics. 2011 Aug;188(4):835-46. doi: 10.1534/genetics.111.128769. Epub 2011 May 30. Genetics. 2011. PMID: 21625001 Free PMC article.
-
Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development.Plant J. 2015 Jul;83(1):78-95. doi: 10.1111/tpj.12830. Epub 2015 Apr 8. Plant J. 2015. PMID: 25781491 Review.
Cited by
-
DNA Repair in Nucleosomes: Insights from Histone Modifications and Mutants.Int J Mol Sci. 2024 Apr 16;25(8):4393. doi: 10.3390/ijms25084393. Int J Mol Sci. 2024. PMID: 38673978 Free PMC article. Review.
-
Unveiling the Conformational Dynamics of the Histone Tails Using Markov State Modeling.J Chem Theory Comput. 2025 May 13;21(9):4921-4938. doi: 10.1021/acs.jctc.5c00196. Epub 2025 Apr 27. J Chem Theory Comput. 2025. PMID: 40289377 Free PMC article.
-
Residues in the Nucleosome Acidic Patch Regulate Histone Occupancy and Are Important for FACT Binding in Saccharomyces cerevisiae.Genetics. 2017 Jul;206(3):1339-1348. doi: 10.1534/genetics.117.201939. Epub 2017 May 3. Genetics. 2017. PMID: 28468903 Free PMC article.
-
Dissecting Nucleosome Function with a Comprehensive Histone H2A and H2B Mutant Library.G3 (Bethesda). 2017 Dec 4;7(12):3857-3866. doi: 10.1534/g3.117.300252. G3 (Bethesda). 2017. PMID: 29038170 Free PMC article.
-
Structural visualization of key steps in nucleosome reorganization by human FACT.Sci Rep. 2019 Jul 15;9(1):10183. doi: 10.1038/s41598-019-46617-7. Sci Rep. 2019. PMID: 31308435 Free PMC article.
References
-
- Gurard-Levin Z.A., Quivy J.P., Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014;83:487–517. - PubMed
-
- Formosa T. Avoiding a fatal attraction: properties of nucleosomes and a histone chaperone revealed under physiological conditions. Mol. Cell. 2010;37:747–748. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

