Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy
- PMID: 27370208
- PMCID: PMC5010046
- DOI: 10.15252/embj.201694491
Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy
Abstract
Mammalian cells deploy autophagy to defend their cytosol against bacterial invaders. Anti-bacterial autophagy relies on the core autophagy machinery, cargo receptors, and "eat-me" signals such as galectin-8 and ubiquitin that label bacteria as autophagy cargo. Anti-bacterial autophagy also requires the kinase TBK1, whose role in autophagy has remained enigmatic. Here we show that recruitment of WIPI2, itself essential for anti-bacterial autophagy, is dependent on the localization of catalytically active TBK1 to the vicinity of cytosolic bacteria. Experimental manipulation of TBK1 recruitment revealed that engagement of TBK1 with any of a variety of Salmonella-associated "eat-me" signals, including host-derived glycans and K48- and K63-linked ubiquitin chains, suffices to restrict bacterial proliferation. Promiscuity in recruiting TBK1 via independent signals may buffer TBK1 functionality from potential bacterial antagonism and thus be of evolutionary advantage to the host.
Keywords: PI(3)P; Salmonella; TBK1; WIPI; anti‐bacterial autophagy.
© 2016 MRC Laboratory of Molecular Biology Published under the terms of the CC BY 4.0 license.
Figures
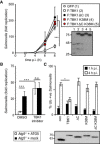
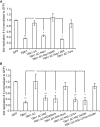
Kinetics of S. Typhimurium replication in Tbk1 −/− MEFs stably expressing the indicated TBK1 alleles. Intracellular bacteria were enumerated by their ability to form colonies on agar plates following cell lysis at the indicated time points. Statistical analysis comparing Tbk1 −/− MEFs and cells expressing TBK1 alleles. Western blot for Flag‐tagged TBK1 variants in post‐nuclear cell lysates.
Replication of S. Typhimurium in Atg5 −/− MEFs complemented with ATG5 or mock and treated with the TBK1 kinase inhibitor MRT68843 (10 nM) or DMSO. Fold replication was determined by counting bacterial colonies on agar plates at 2 and 8 h post‐inoculation (p.i.) following cell lysis.
Analysis of Tbk1 −/− MEFs stably expressing the indicated TBK1 alleles and infected with S. Typhimurium. Percentage of S. Typhimurium coated with ubiquitin (detected by FK2 antibody) at 1 or 4 h p.i. Western blot for Flag‐tagged TBK1 variants in post‐nuclear cell lysates.
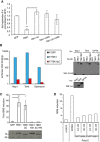
Replication of Salmonella enterica serovar Typhimurium in Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1ΔC fusion proteins. Replication normalized to cells expressing GFP. Mean and SEM. N = 3, ***P < 0.001 one‐way ANOVA with Dunnett's multiple comparisons test.
Lumier binding assay. Binding of luciferase‐tagged Nap1, Tank, or optineurin to beads coated with Flag:GFP, Flag:TBK1, or Flag:TBK1ΔC. Western blot for luciferase‐tagged or Flag‐tagged proteins in total cell lysates.
ISRE reporter induction upon transient expression of the indicated Flag:TBK1 alleles into 293ET cells. Western blot for Flag‐tagged proteins. Mean and SEM of four independent experiments. *P < 0.05, **P < 0.01, Student's t‐test.
ISRE reporter induction upon stimulation of 293ET cells with polyI:C (8 h) in the presence of DMSO or TBK1 inhibitors MRT68843 or MRT68601.

- A–H
Analysis of Tbk1 −/− MEFs stably expressing the indicated TBK1 alleles and infected with S. Typhimurium for 1 h. Percentage of S. Typhimurium coated with GFP:LC3B (A), the indicated GFP:WIPI alleles (B), GFP:DFCP1 (FYVE* denotes a PI(3)P‐binding mutant of DFCP1) (D, E), or GFP:ML1N*2, a probe for PI(3,5)P2 (G). Where indicated, wortmannin (Wort) was added at 100 nM. Confocal micrographs of MEFs expressing the indicated GFP fusion proteins or immunolabeled for WIPI2 and infected with mCherry‐expressing S. Typhimurium (C, F and H). Mean and SEM of at least three independent experiments with duplicate coverslips. > 200 bacteria counted per coverslip. *P < 0.05, **P < 0.01 one‐way ANOVA with Dunnett's multiple comparisons test. Scale bar, 10 μm.
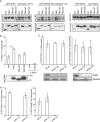
- A
Western blots of GFP‐tagged WIPI alleles and Flag‐tagged TBK1 alleles corresponding to Fig 2B.
- B
Percentage of WIPI2‐positive S. Typhimurium at 1 h p.i., in Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1 proteins. Wortmannin (Wort) was added as indicated at 100 nM. Mean and SEM. N = 3, *P < 0.05, one‐way ANOVA with Dunnett's multiple comparisons test. Western blot of TBK1 alleles in post‐nuclear lysates.
- C, D
Analysis of MEFs, stably expressing GFP:WIPI1 or GFP:WIPI2B, treated with the indicated siRNAs and infected with S. Typhimurium for 1 h. Mean and SEM. N = 3. Western blot for optineurin and tubulin upon the indicated siRNA treatment.
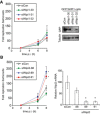
- A, B
Kinetics of Salmonella enterica serovar Typhimurium replication in MEFs depleted of WIPI1 (A) or WIPI2 (B). Intracellular bacteria were enumerated by their ability to form colonies on agar plates. Western blot for GFP:WIPI1 and quantitative RT–PCR for WIPI2 upon the indicated siRNA treatments. Mean and SEM. N = 6 (A), n = 3 (B). *P < 0.05, Student's t‐test.

- A–D
Analysis of Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1ΔC‐adaptor fusion proteins. S. Typhimurium replication kinetics (A, D). Infected cells were lysed at the indicated time points post‐inoculation (p.i.), and bacteria were enumerated by their ability to form colonies on agar plates. Mean and SD of triplicate MEF cultures and duplicate colony counts. Data are representative of at least two repeats. Statistical significance to Tbk1 −/− MEFs expressing TBK1ΔC is indicated. Western blot for Flag‐tagged TBK1 variants in post‐nuclear cell lysates. (B) Percentage of GFP:WIPI1‐positive S. Typhimurium at 1 h p.i. in the indicated complemented MEFs. Mean and SEM of four independent experiments. > 200 bacteria counted per coverslip. *P < 0.05, ***P < 0.001, one‐way ANOVA with Dunnett's multiple comparisons test. (C) TBK1–Nap1–NDP52 complex. N‐ and C‐termini, domains and binding partners are indicated.
- A, B
Replication of S. Typhimurium in Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1ΔC alleles. Replication normalized to cells expressing GFP. Mean and SEM. N = 3, *P < 0.05, **P < 0.01, one‐way ANOVA with Dunnett's multiple comparisons test.
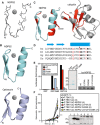
Superposition of the 20 lowest energy solution structures of the NDP52 domain
Cartoon representation of the zinc finger domains of NDP52 and optineurin (pdb id: 5AAZ)
Cartoon representation of the NDP52 zinc finger and ubiquitin. In red are those residues whose signals alter upon binding.
Phylogenetic alignment of the NDP52 zinc finger. Zinc‐coordinating residues in bold, conserved residues in the helix in red. The asterisk indicates D439, the residue mutated in subsequent experiments. Hs: Homo sapiens, Bt: Bos taurus, Gg: Gallus gallus, and Xe: Xenopus laevis.
Lumier binding assay. Binding of the indicated luciferase‐tagged NDP52 variants to beads coated with GST, GST:galectin‐8, or GST:ubiquitin. Coomassie stain of purified GST proteins. Western blot for luciferase‐tagged NDP52 alleles in total cell lysates.
Kinetics of Salmonella enterica serovar Typhimurium replication in Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1ΔC‐NDP52 fusion proteins. Infected cells were lysed at the indicated times post‐inoculation (p.i.) and bacteria were enumerated by their ability to form colonies on agar plates. Mean and SD of triplicate MEF cultures and duplicate colony counts. Data are representative of at least two repeats. Statistical comparison with Tbk1 −/− MEFs expressing TBK1ΔC is indicated. ***P < 0.001, one‐way ANOVA with Dunnett's multiple comparisons test. Western blot for Flag‐tagged TBK1 variants in post‐nuclear cell lysates.

- A, B
Analysis of Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1ΔC‐galectin fusion proteins. S. Typhimurium replication kinetics (A). Infected cells were lysed at the indicated times post‐inoculation (p.i.), and bacteria were enumerated by their ability to form colonies on agar plates. Mean and SD of triplicate MEF cultures and duplicate colony counts. Data are representative of at least two repeats. Statistical differences to Tbk1 −/− MEFs expressing TBK1ΔC are shown. ***P < 0.001, one‐way ANOVA with Dunnett's multiple comparisons test. Western blot for Flag‐tagged TBK1 variants in post‐nuclear cell lysates. (B) Percentage of GFP:WIPI1‐positive S. Typhimurium at 1 h p.i. in the indicated complemented MEFs. Mean and SEM of three independent experiments. > 200 bacteria counted per coverslip. *P < 0.05, one‐way ANOVA with Dunnett's multiple comparisons test.
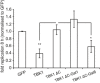

- A
Binding of NDP52 to ubiquitin tetramers of different linkage types. Beads coated with the indicated GST‐tagged proteins were incubated with pooled M1‐, K63‐, and K48‐linked tetra‐ubiquitin chains. Anti‐ubiquitin Western blot of input and proteins bound to beads.
- B–D
Kinetics of S. Typhimurium replication in Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1ΔC fusion proteins. Infected cells were lysed at the indicated time points post‐inoculation (p.i.) and bacteria were enumerated by their ability to form colonies on agar plates. Mean and SD of triplicate MEF cultures and duplicate colony counts. Data are representative of at least two repeats. Statistical differences to Tbk1 −/− MEFs expressing TBK1ΔC are shown. *P < 0.5, **P < 0.01, ***P < 0.001, one‐way ANOVA with Dunnett's multiple comparisons test. Western blot for Flag‐tagged TBK1 variants in post‐nuclear cell lysates.

- A–C
Replication of S. Typhimurium in Tbk1 −/− MEFs complemented with the indicated Flag‐tagged TBK1ΔC fusion proteins. Replication normalized to cells expressing GFP. Mean and SEM. N = 4, *P < 0.05, **P < 0.01, ***P < 0.001, one‐way ANOVA with Dunnett's multiple comparisons test.
References
-
- Boyle KB, Randow F (2013) The role of “eat‐me” signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol 16: 1–10 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

