Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
- PMID: 27371726
- PMCID: PMC4976021
- DOI: 10.4049/jimmunol.1600589
Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Abstract
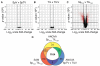
Mechanisms of self-tolerance often result in CD8(+) tumor-infiltrating lymphocytes (TIL) with a hypofunctional phenotype incapable of tumor clearance. Using a transplantable colon carcinoma model, we found that CD8(+) T cells became tolerized in <24 h in an established tumor environment. To define the collective impact of pathways suppressing TIL function, we compared genome-wide mRNA expression of tumor-specific CD8(+) T cells from the tumor and periphery. Notably, gene expression induced during TIL hypofunction more closely resembled self-tolerance than viral exhaustion. Differential gene expression was refined to identify a core set of genes that defined hypofunctional TIL; these data comprise the first molecular profile of tumor-specific TIL that are naturally responding and represent a polyclonal repertoire. The molecular profile of TIL was further dissected to determine the extent of overlap and distinction between pathways that collectively restrict T cell functions. As suggested by the molecular profile of TIL, protein expression of inhibitory receptor LAG-3 was differentially regulated throughout prolonged late-G1/early-S phase of the cell cycle. Our data may accelerate efficient identification of combination therapies to boost anti-tumor function of TIL specifically against tumor cells.
Copyright © 2016 by The American Association of Immunologists, Inc.
Figures
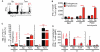




References
-
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. - PubMed
-
- Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. - PubMed
-
- Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. - PubMed
-
- Baitsch L, Fuertes-Marraco SA, Legat A, Meyer C, Speiser DE. The three main stumbling blocks for anticancer T cells. Trends Immunol. 2012;33:364–372. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

