2.8-Å Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania
- PMID: 27373148
- PMCID: PMC5835689
- DOI: 10.1016/j.celrep.2016.06.014
2.8-Å Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania
Abstract
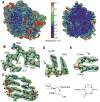
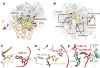
Leishmania is a single-cell eukaryotic parasite of the Trypanosomatidae family, whose members cause an array of tropical diseases. The often fatal outcome of infections, lack of effective vaccines, limited selection of therapeutic drugs, and emerging resistant strains, underline the need to develop strategies to combat these pathogens. The Trypanosomatid ribosome has recently been highlighted as a promising therapeutic target due to structural features that are distinct from other eukaryotes. Here, we present the 2.8-Å resolution structure of the Leishmania donovani large ribosomal subunit (LSU) derived from a cryo-EM map, further enabling the structural observation of eukaryotic rRNA modifications that play a significant role in ribosome assembly and function. The structure illustrates the unique fragmented nature of leishmanial LSU rRNA and highlights the irregular distribution of rRNA modifications in Leishmania, a characteristic with implications for anti-parasitic drug development.
Copyright © 2016. Published by Elsevier Inc.
Figures


References
-
- Afonine PV, Headd JJ, Terwilliger TC, Adams PD. New tool: phenix.real_space_refine. Computational Crystallography Newsletter. 2013;4:43–44.
-
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. - PubMed
-
- Eliaz D, Doniger T, Tkacz ID, Biswas VK, Gupta SK, Kolev NG, Unger R, Ullu E, Tschudi C, Michaeli S, et al. Genome-wide analysis of small nucleolar RNAs of Leishmania major reveals a rich repertoire of RNAs involved in modification and processing of rRNA. RNA biology. 2015;12:1222–1255. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

