Transcription Factor NFIB Is a Driver of Small Cell Lung Cancer Progression in Mice and Marks Metastatic Disease in Patients
- PMID: 27373156
- PMCID: PMC4956617
- DOI: 10.1016/j.celrep.2016.06.020
Transcription Factor NFIB Is a Driver of Small Cell Lung Cancer Progression in Mice and Marks Metastatic Disease in Patients
Abstract
Small cell lung cancer (SCLC) is an aggressive neuroendocrine tumor, and no effective treatment is available to date. Mouse models of SCLC based on the inactivation of Rb1 and Trp53 show frequent amplifications of the Nfib and Mycl genes. Here, we report that, although overexpression of either transcription factor accelerates tumor growth, NFIB specifically promotes metastatic spread. High NFIB levels are associated with expansive growth of a poorly differentiated and almost exclusively E-cadherin (CDH1)-negative invasive tumor cell population. Consistent with the mouse data, we find that NFIB is overexpressed in almost all tested human metastatic high-grade neuroendocrine lung tumors, warranting further assessment of NFIB as a tumor progression marker in a clinical setting.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

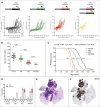
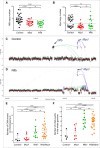
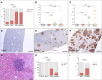


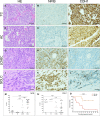
Comment in
-
Recalcitrant small cell lung cancer: the argument for optimism.J Thorac Dis. 2017 Mar;9(3):E295-E296. doi: 10.21037/jtd.2017.03.40. J Thorac Dis. 2017. PMID: 28449525 Free PMC article. No abstract available.
References
-
- Andreasen S., Persson M., Kiss K., Homøe P., Heegaard S., Stenman G. Genomic profiling of a combined large cell neuroendocrine carcinoma of the submandibular gland. Oncol. Rep. 2016;35:2177–2182. - PubMed
-
- Calbo J., van Montfort E., Proost N., van Drunen E., Beverloo H.B., Meuwissen R., Berns A. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19:244–256. - PubMed
-
- Cermáková K., Tesina P., Demeulemeester J., El Ashkar S., Méreau H., Schwaller J., Rezáčová P., Veverka V., De Rijck J. Validation and structural characterization of the LEDGF/p75-MLL interface as a new target for the treatment of MLL-dependent leukemia. Cancer Res. 2014;74:5139–5151. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

